
Burak Avcı on Twitter: "(4/8) This solid 16S rRNA sequence database enabled us to design specific oligonucleotide probes to visualise Loki- and Heimdallarchaeota cells using catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH).

New ribosomal RNA BLAST databases available on the web BLAST service and for download - NCBI Insights
![Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis [PeerJ] Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/10778/1/fig-1-full.png)
Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis [PeerJ]
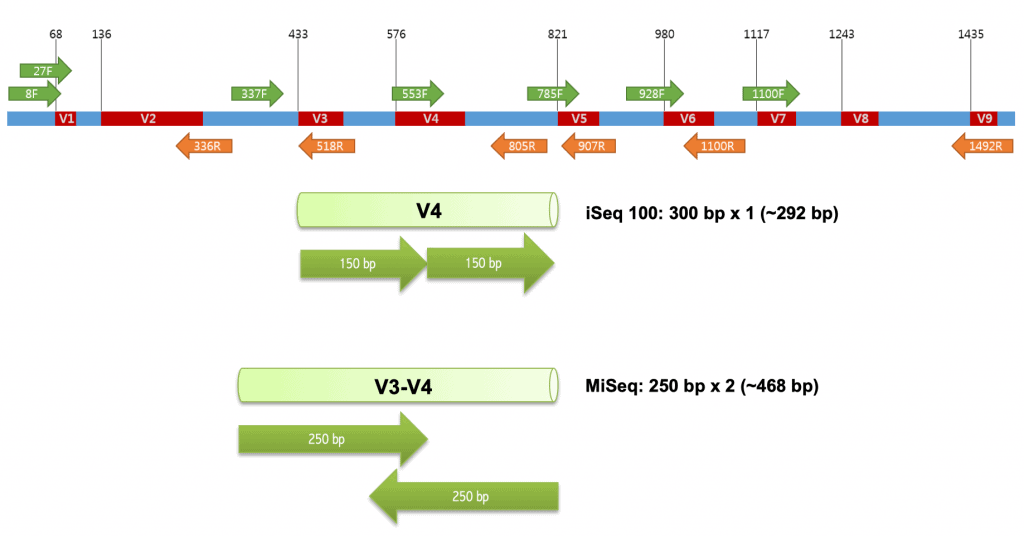
Soil Bacterial Diversity Screening Using Single 16S rRNA Gene V Regions Coupled with Multi-Million Read Generating Sequencing Technologies | PLOS ONE

The workflow of this study. (A) The taxonomy of 16S rRNA sequences in... | Download Scientific Diagram

Full‐length 16S rRNA gene classification of Atlantic salmon bacteria and effects of using different 16S variable regions on community structure analysis - Klemetsen - 2019 - MicrobiologyOpen - Wiley Online Library

IJMS | Free Full-Text | Targeting the 16S rRNA Gene for Bacterial Identification in Complex Mixed Samples: Comparative Evaluation of Second (Illumina) and Third (Oxford Nanopore Technologies) Generation Sequencing Technologies
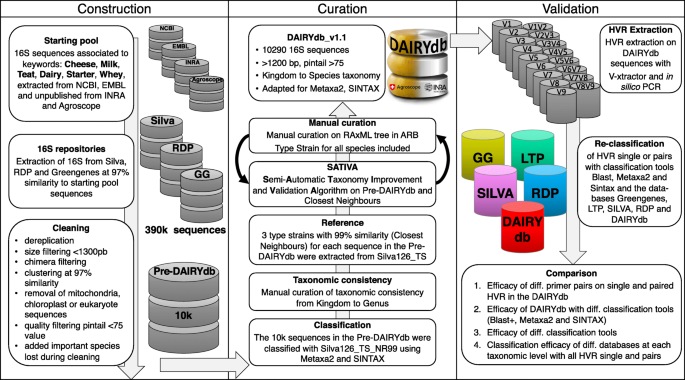
DAIRYdb: a manually curated reference database for improved taxonomy annotation of 16S rRNA gene sequences from dairy products | BMC Genomics | Full Text
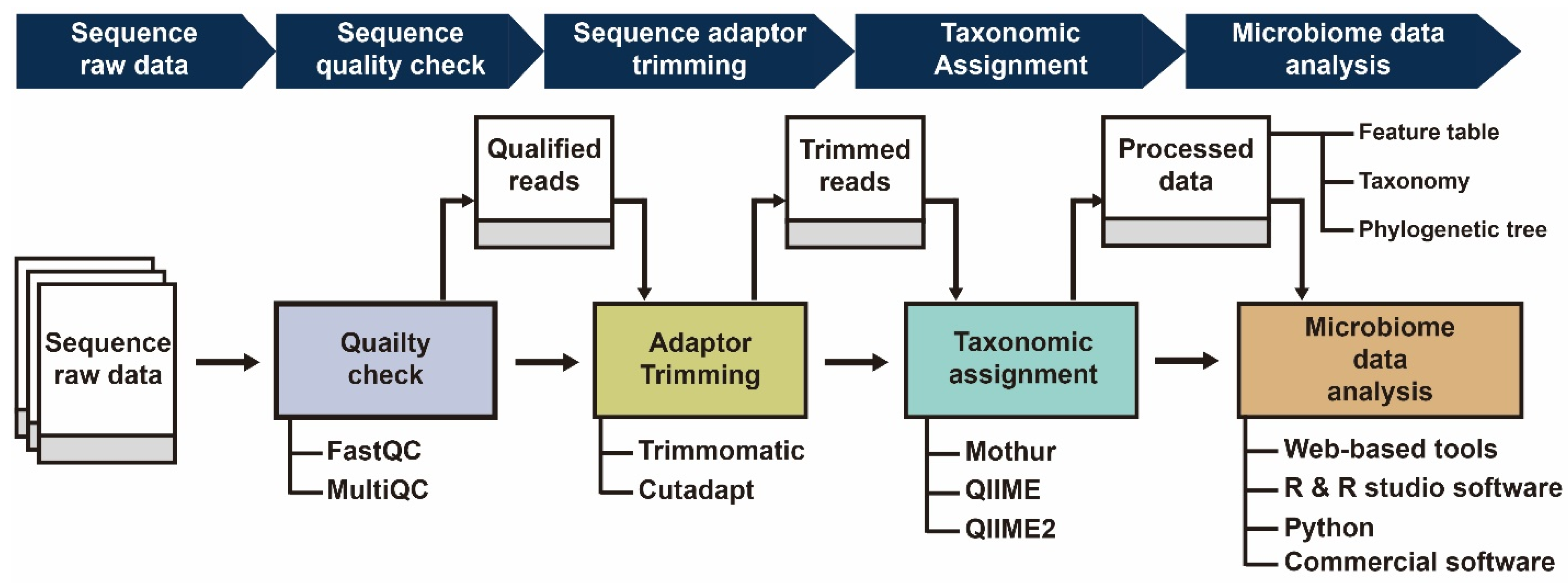
IJMS | Free Full-Text | Review of the Current State of Freely Accessible Web Tools for the Analysis of 16S rRNA Sequencing of the Gut Microbiome

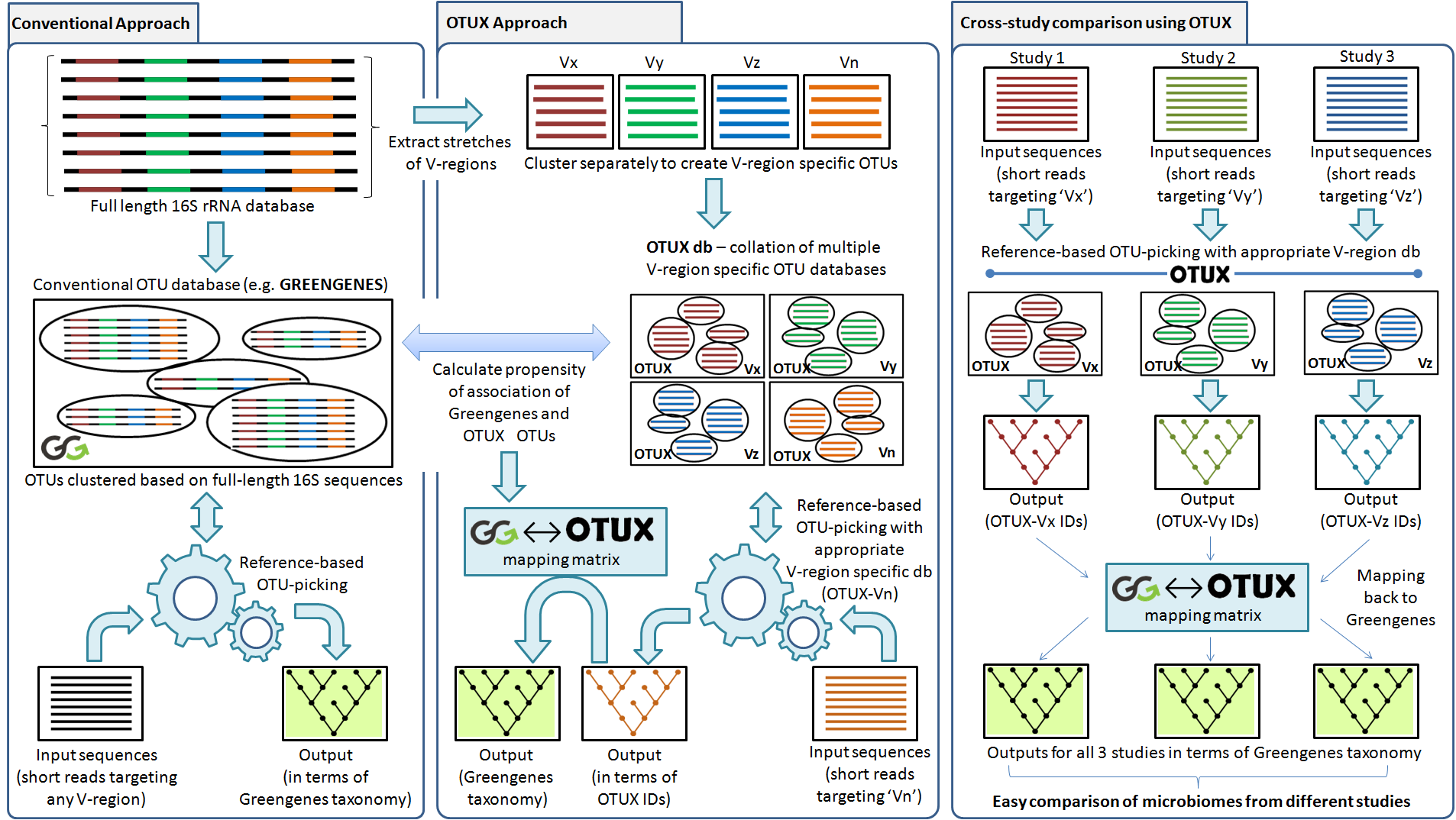
Construction & assessment of a unified curated reference database for improving the taxonomic classification of bacteria using 16S rRNA sequence data | Semantic Scholar
![PDF] Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies | Semantic Scholar PDF] Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/2033072bcbe5655b243700a0a65134e9dc1b58d0/8-Figure1-1.png)
PDF] Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies | Semantic Scholar
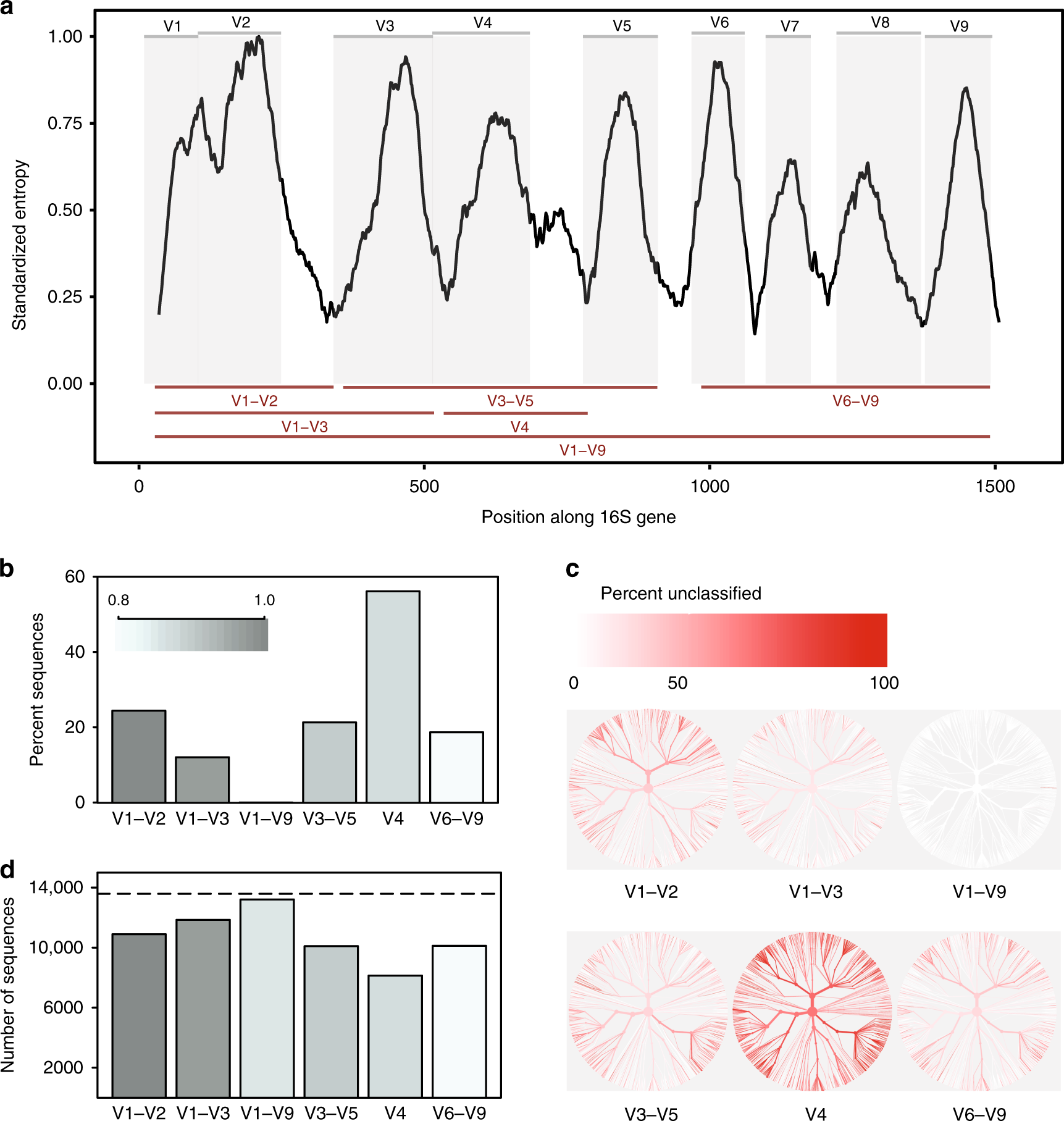
Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis | Nature Communications
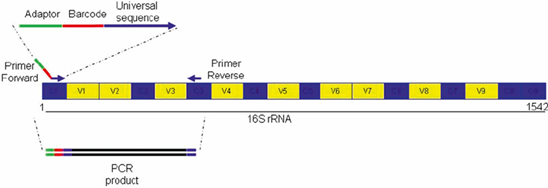
16s rRNA Sequencing Analysis | Microbiome Sequencing – 1010Genome | Quality NGS Bioinformatics Data Analysis Services
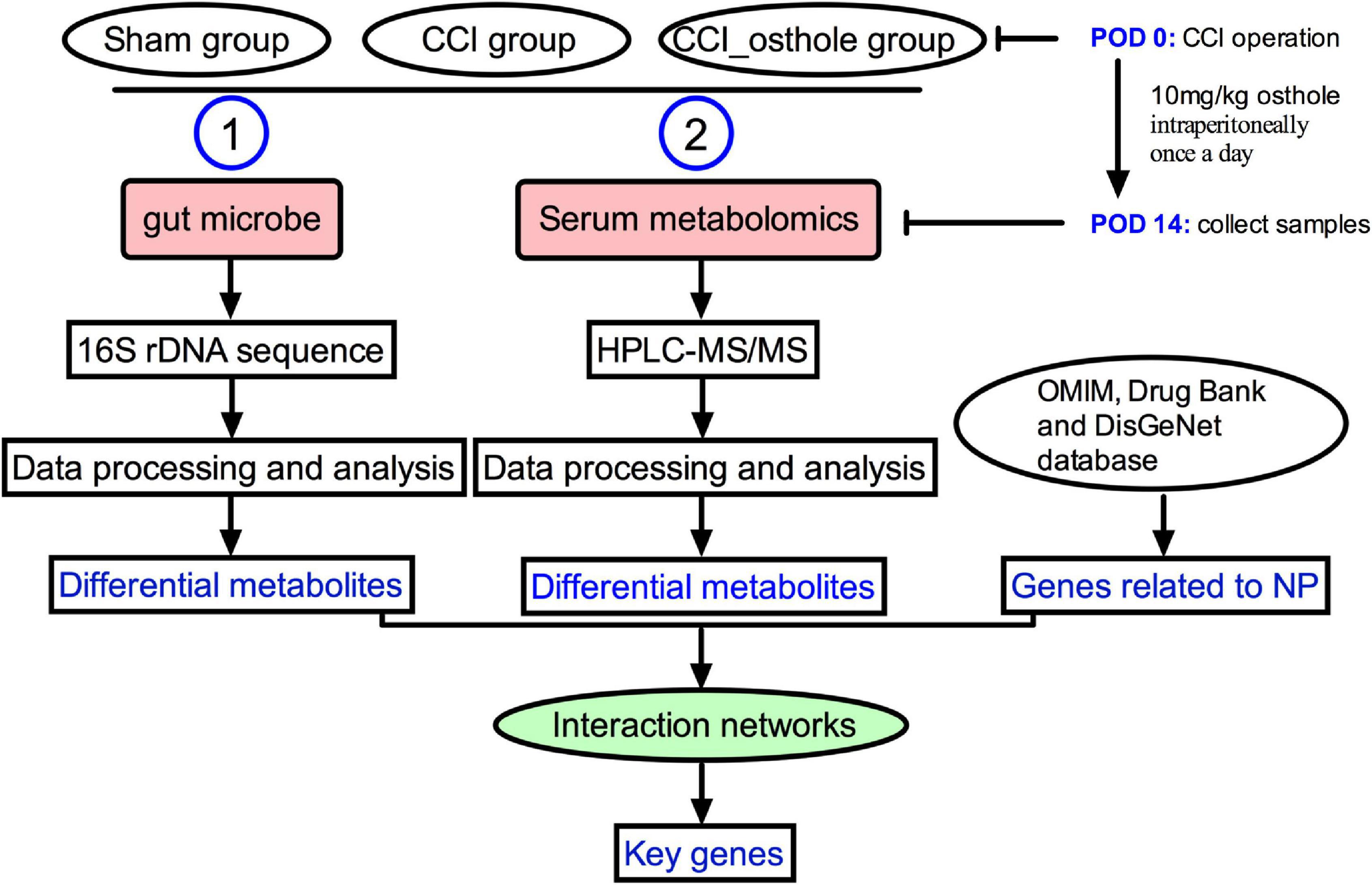
Frontiers | Integrated 16S rRNA Gene Sequencing and Metabolomics Analysis to Investigate the Important Role of Osthole on Gut Microbiota and Serum Metabolites in Neuropathic Pain Mice



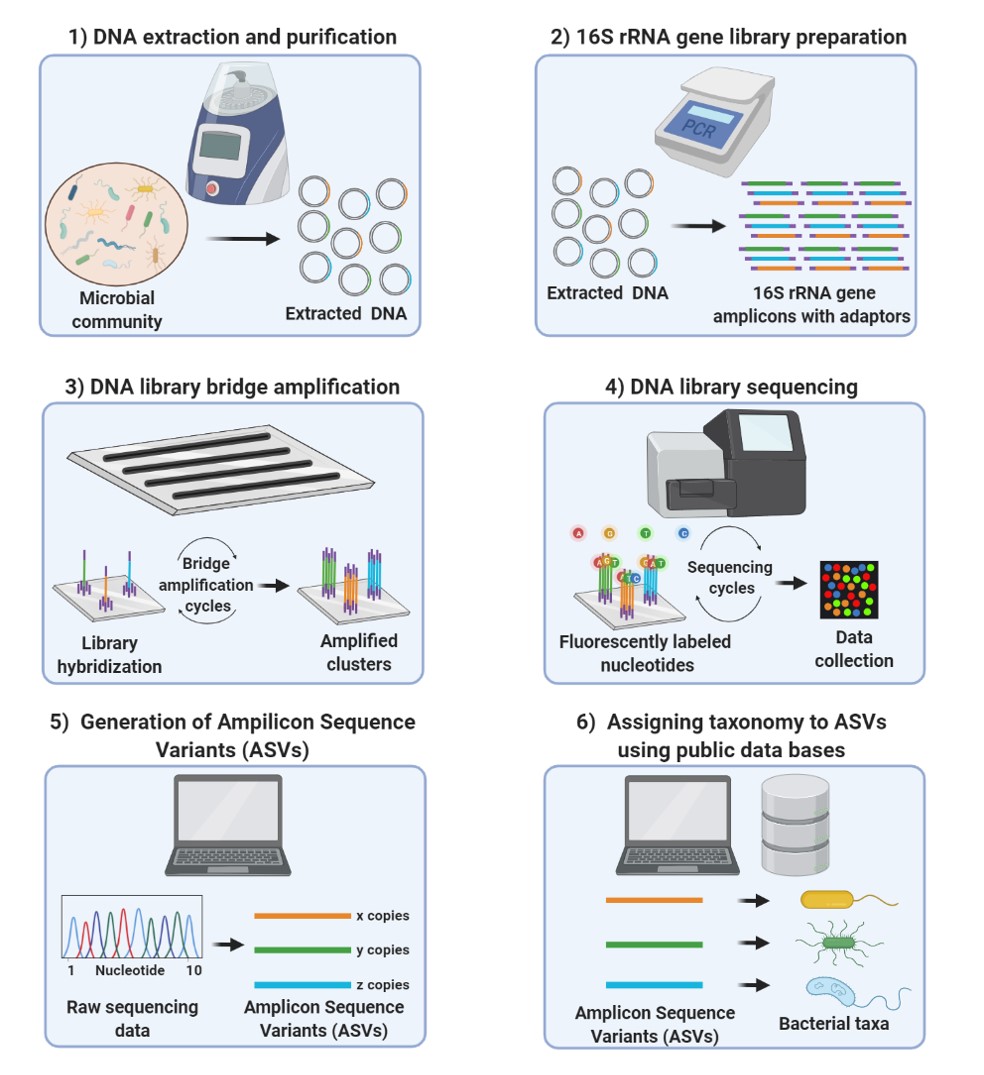



![De novo species identification using 16S rRNA gene nanopore sequencing [PeerJ] De novo species identification using 16S rRNA gene nanopore sequencing [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2020/10029/1/fig-1-2x.jpg)