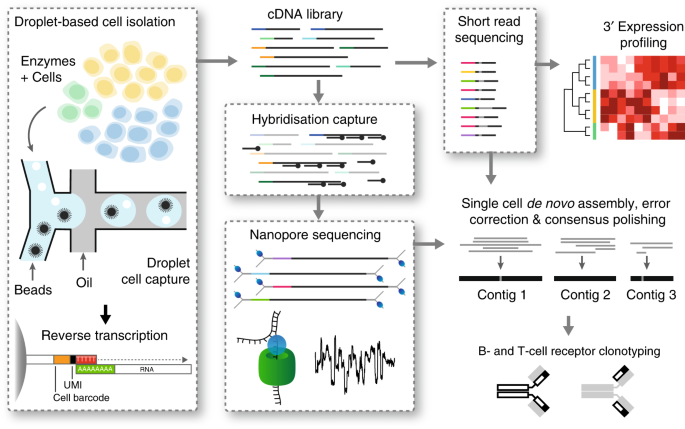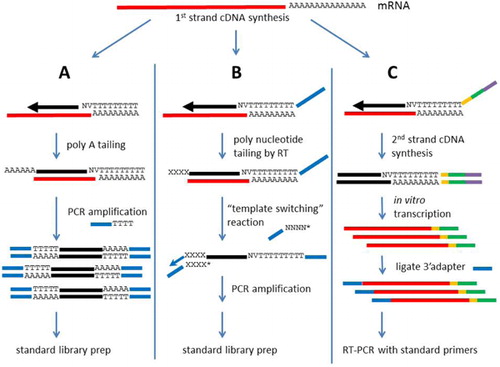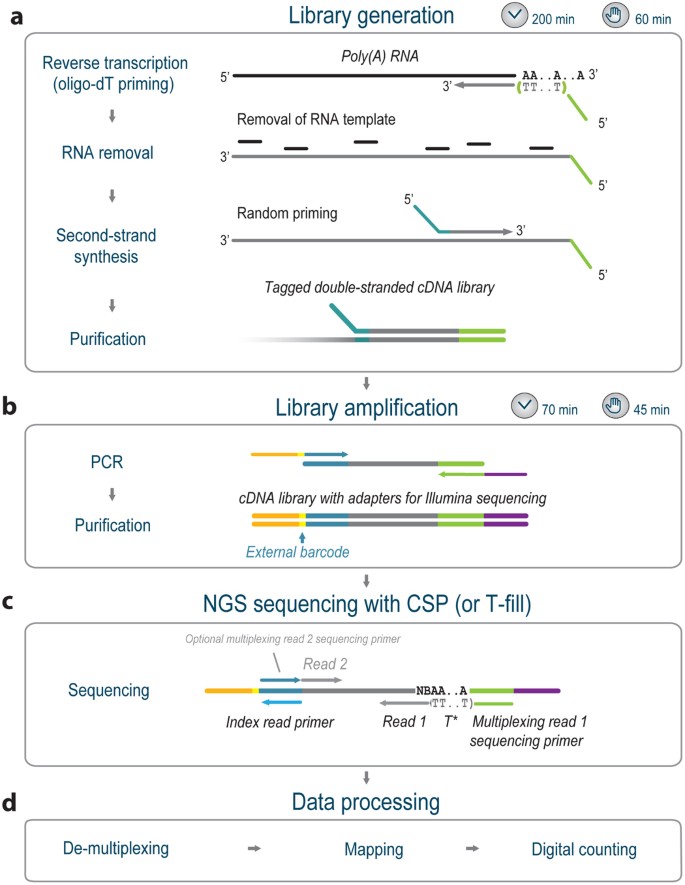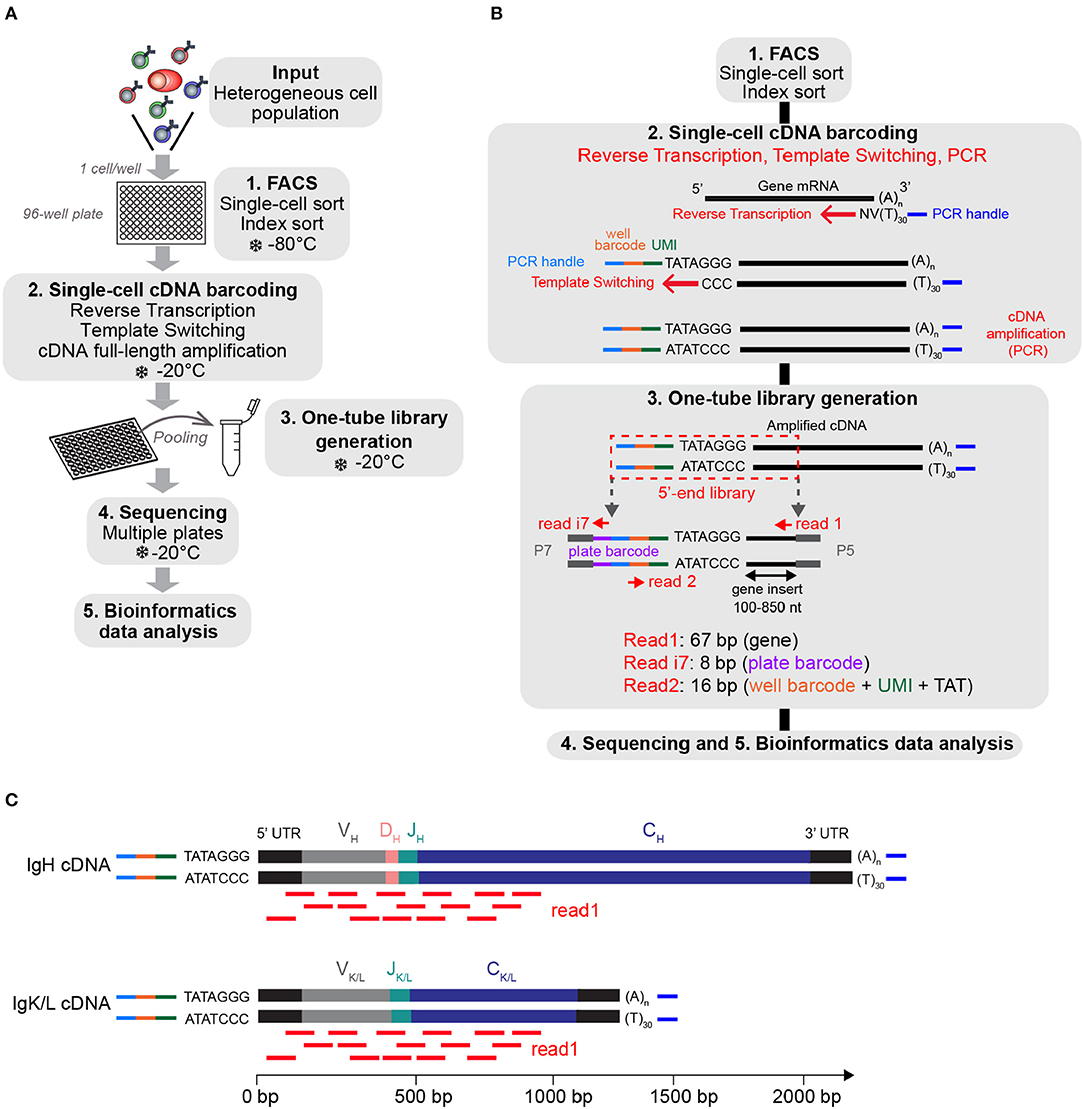
Frontiers | FB5P-seq: FACS-Based 5-Prime End Single-Cell RNA-seq for Integrative Analysis of Transcriptome and Antigen Receptor Repertoire in B and T Cells
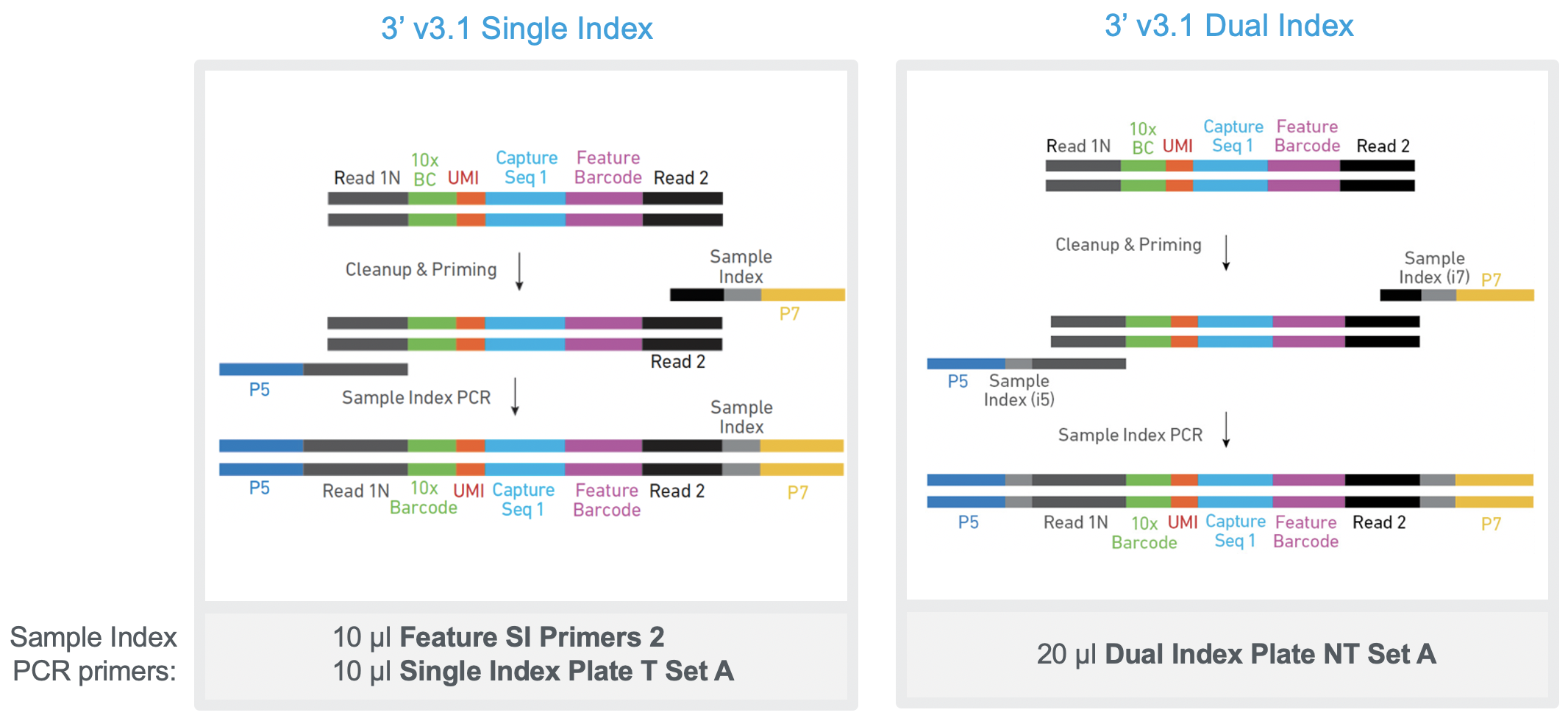
What are the protocol differences between Single Cell 3' v3.1 Single Index and 3' v3.1 Dual Index? – 10X Genomics
GitHub - Reda94/QuantSeq-3-prime-FWD-RNAseq-pipeline: Implementation of the RNA-Seq analysis pipeline used on sequencing data obtained from Lexogen's QuantSeq FWD 3' mRNA-Seq kit.
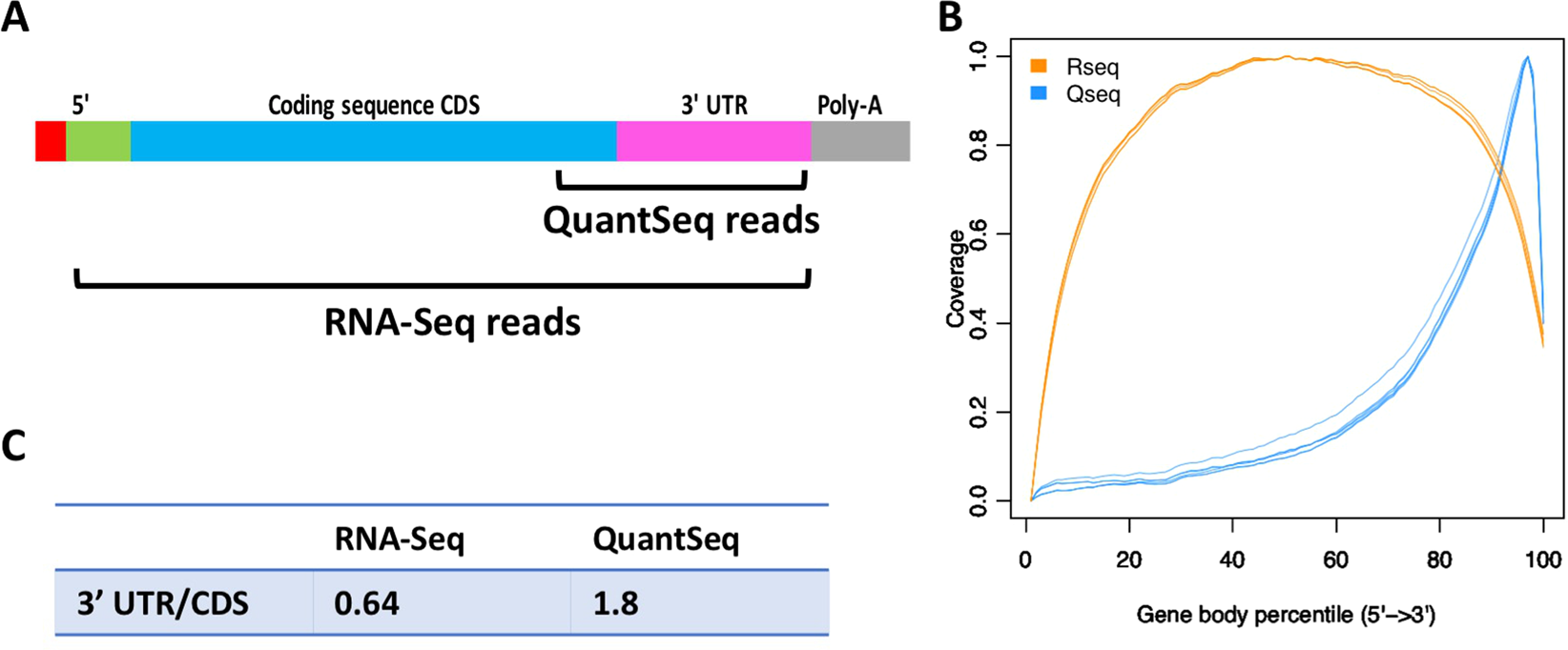
QuantSeq. 3′ Sequencing combined with Salmon provides a fast, reliable approach for high throughput RNA expression analysis | Scientific Reports
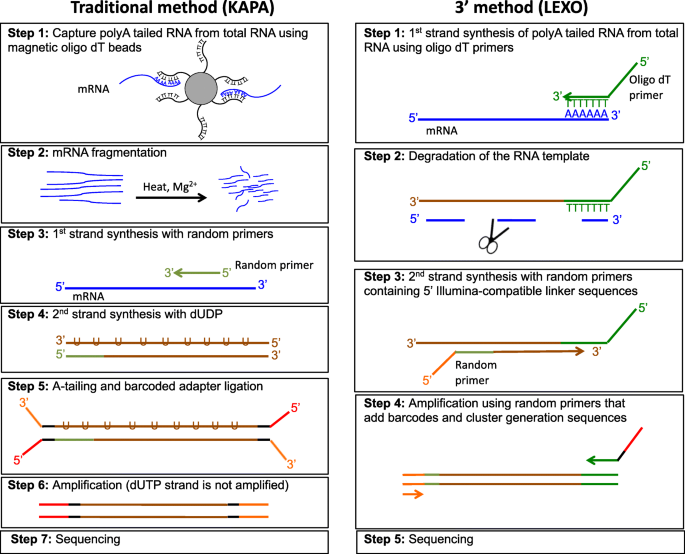
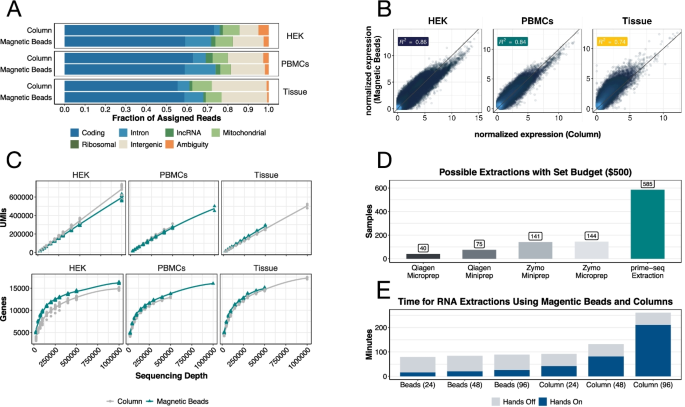
A comparison between whole transcript and 3' RNA sequencing methods using Kapa and Lexogen library preparation methods | BMC Genomics | Full Text

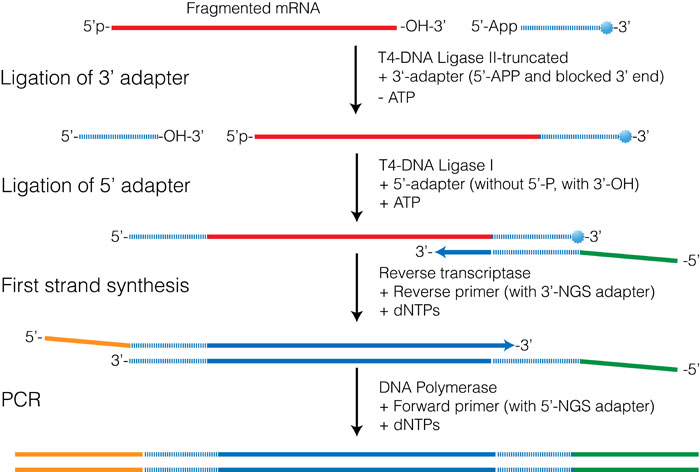
3′ RACE with deep sequencing, schematic of method used. a Ligation of... | Download Scientific Diagram