
The Shine-Dalgarno sequence of riboswitch-regulated single mRNAs shows ligand-dependent accessibility bursts | Nature Communications

Elucidating the 16S rRNA 3′ boundaries and defining optimal SD/aSD pairing in Escherichia coli and Bacillus subtilis using RNA-Seq data | Scientific Reports
Frontiers | Comparative Analysis of anti-Shine- Dalgarno Function in Flavobacterium johnsoniae and Escherichia coli

A Snapshot of the 30S Ribosomal Subunit Capturing mRNA via the Shine- Dalgarno Interaction: Structure
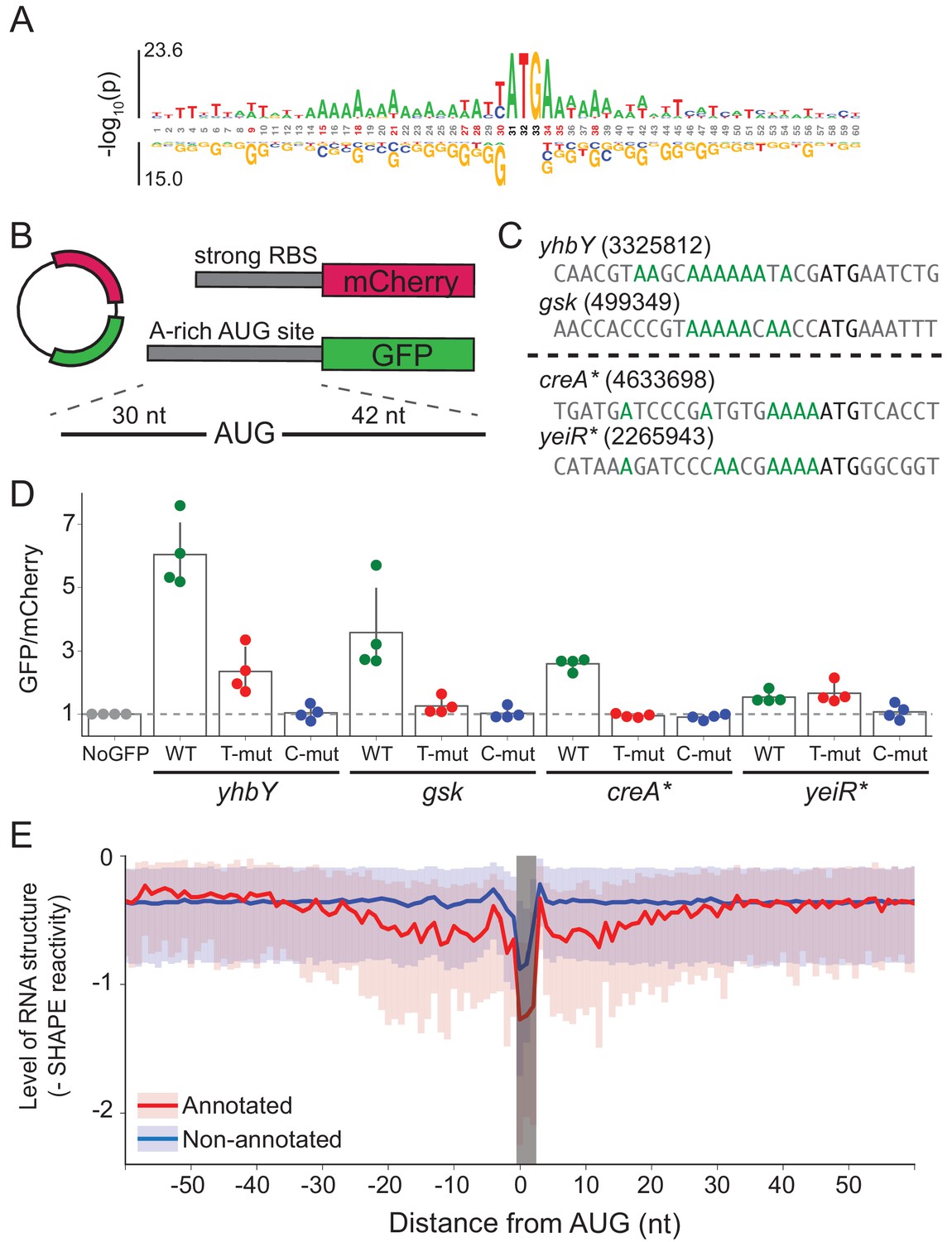
Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing | eLife

Sequence entropy quantifies genome-wide SD sequence utilization. (A),... | Download Scientific Diagram
What do biochemistry students pay attention to in external representations of protein translation? The case of the Shine

What is the Difference Between Shine Dalgarno and Kozak Sequence | Compare the Difference Between Similar Terms
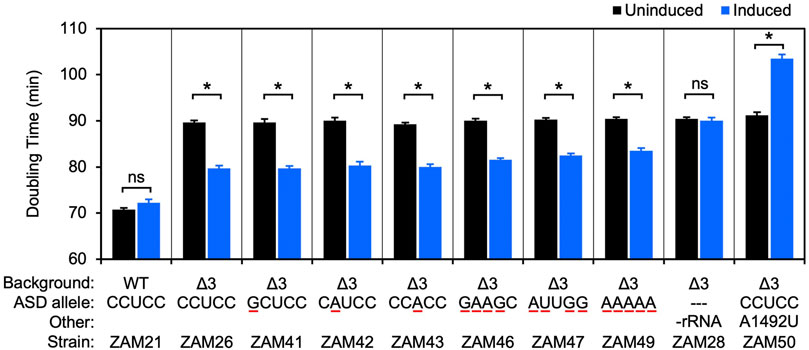
Defining the anti-Shine-Dalgarno sequence interaction and quantifying its functional role in regulating translation efficiency
![PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/ca6e3ce301334ffc5d3a7df6b8a8e5d2bc19ed16/23-Figure1.2-1.png)
PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar

Extending the Spacing between the Shine–Dalgarno Sequence and P-Site Codon Reduces the Rate of mRNA Translocation - ScienceDirect

Accessibility of the Shine-Dalgarno Sequence Dictates N-Terminal Codon Bias in E. coli - ScienceDirect
Re-annotation of 12,495 prokaryotic 16S rRNA 3' ends and analysis of Shine- Dalgarno and anti-Shine-Dalgarno sequences | PLOS ONE

Influences on gene expression in vivo by a Shine–Dalgarno sequence - Jin - 2006 - Molecular Microbiology - Wiley Online Library

Leveraging genome-wide datasets to quantify the functional role of the anti- Shine–Dalgarno sequence in regulating translation efficiency | Open Biology
Leveraging genome-wide datasets to quantify the functional role of the anti- Shine–Dalgarno sequence in regulating translation efficiency - Publications - Amaral Lab

An extended Shine–Dalgarno sequence in mRNA functionally bypasses a vital defect in initiator tRNA | PNAS







