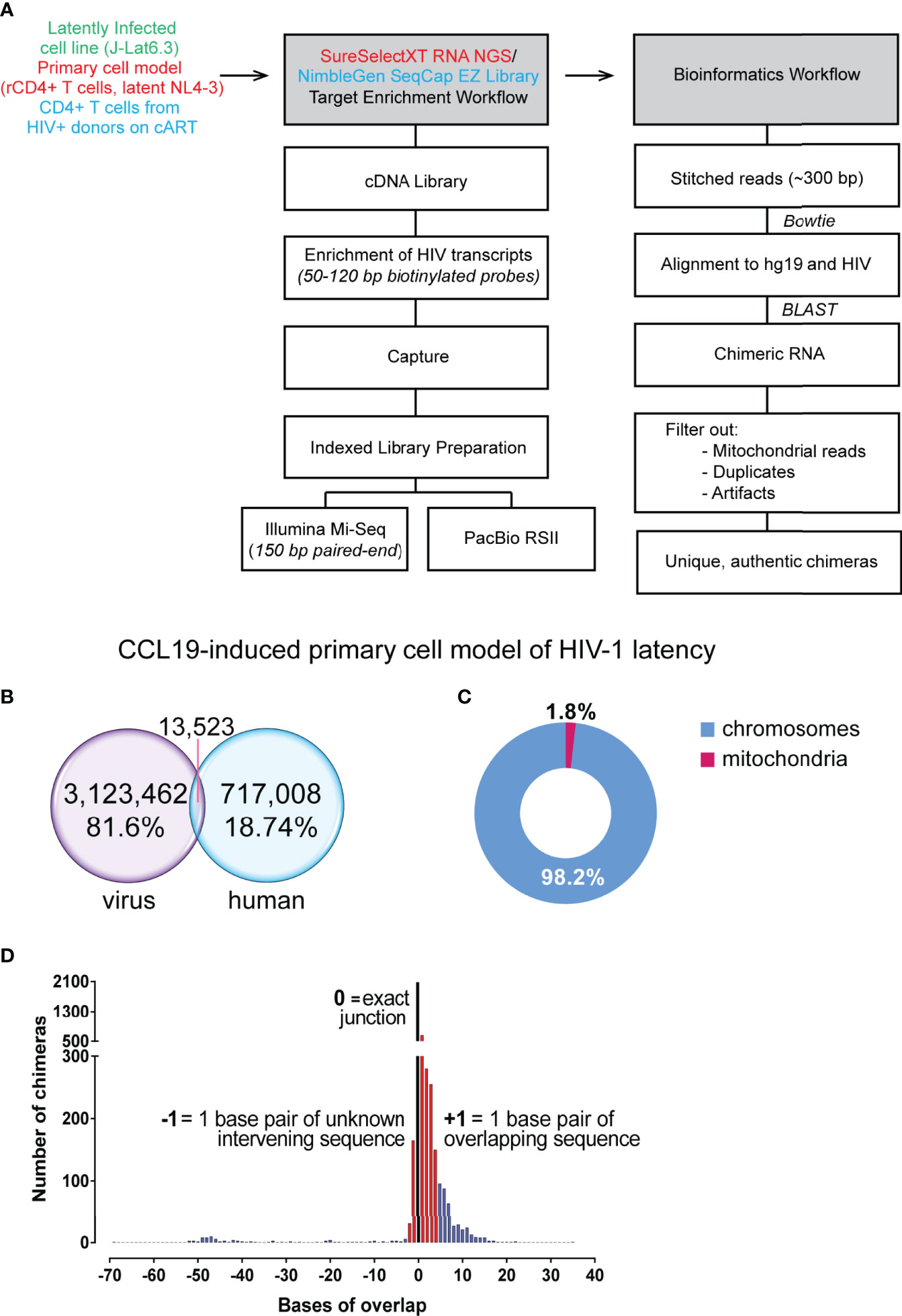
Frontiers | Detection of Chimeric Cellular: HIV mRNAs Generated Through Aberrant Splicing in HIV-1 Latently Infected Resting CD4+ T Cells

Recovery of microbial community profile information hidden in chimeric sequence reads - ScienceDirect

An example of microhomology-induced chimeric read (MICR)-originated... | Download Scientific Diagram

Widespread Transcriptional Readthrough Caused by Nab2 Depletion Leads to Chimeric Transcripts with Retained Introns - ScienceDirect

DECIPHER, a Search-Based Approach to Chimera Identification for 16S rRNA Sequences | Applied and Environmental Microbiology
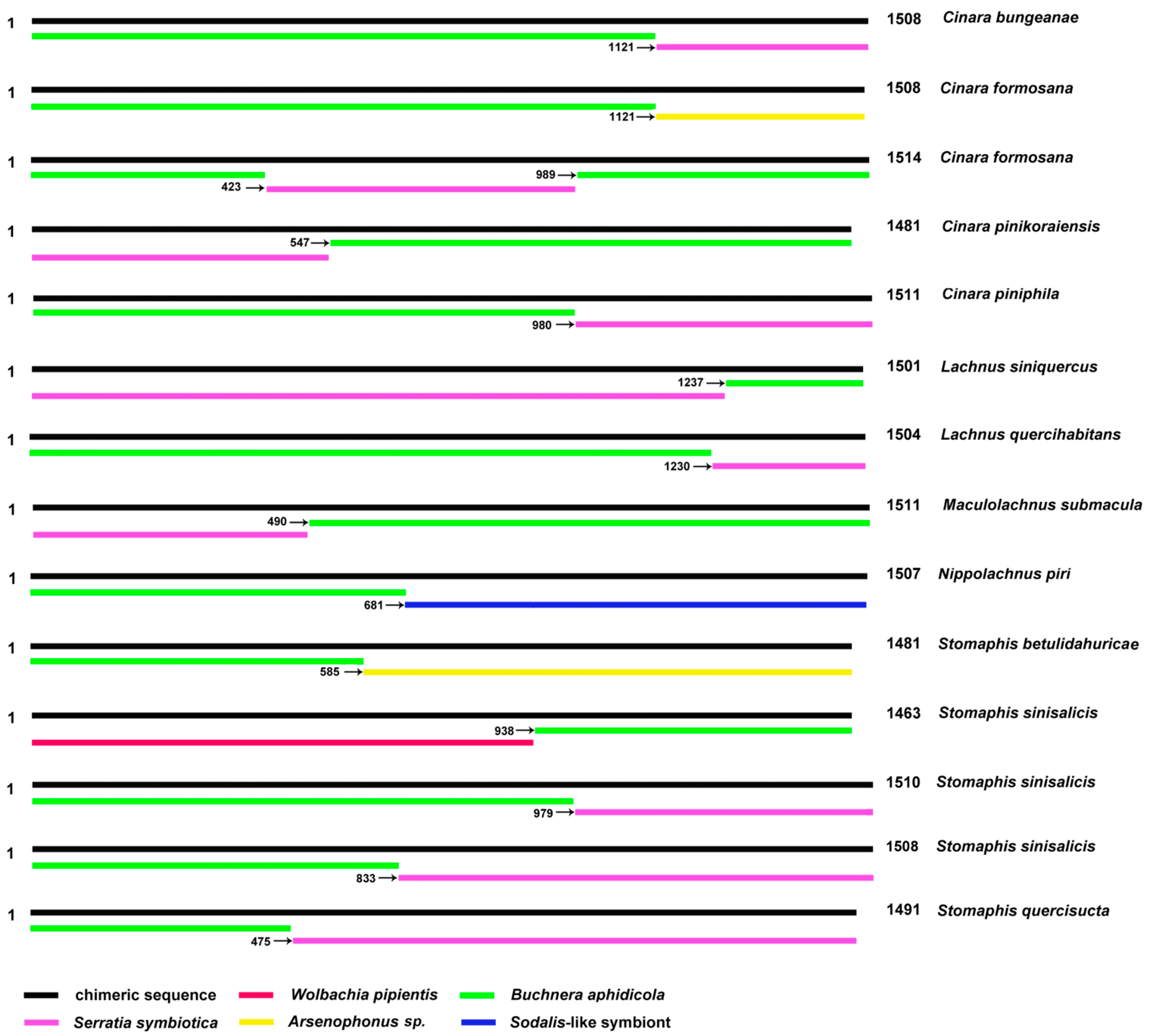
IJMS | Free Full-Text | Avoidance and Potential Remedy Solutions of Chimeras in Reconstructing the Phylogeny of Aphids Using the 16S rRNA Gene of Buchnera: A Case in Lachninae (Hemiptera)

IJMS | Free Full-Text | Avoidance and Potential Remedy Solutions of Chimeras in Reconstructing the Phylogeny of Aphids Using the 16S rRNA Gene of Buchnera: A Case in Lachninae (Hemiptera)

Identification and quantification of chimeric sequencing reads in a highly multiplexed RAD-seq protocol | bioRxiv
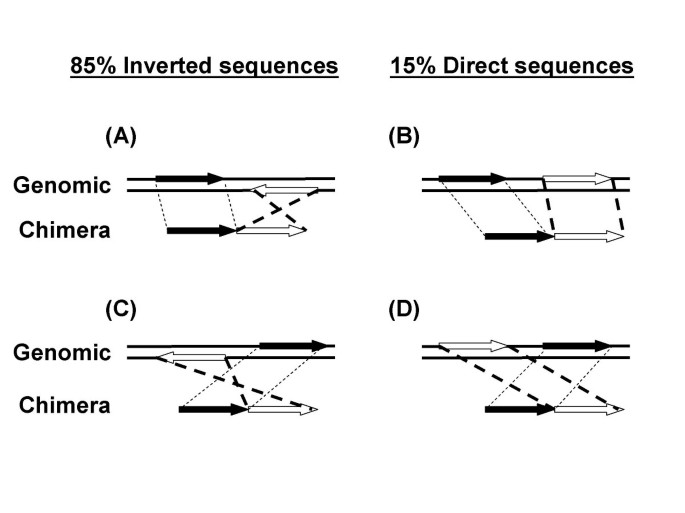
Mechanism of chimera formation during the Multiple Displacement Amplification reaction | BMC Biotechnology | Full Text
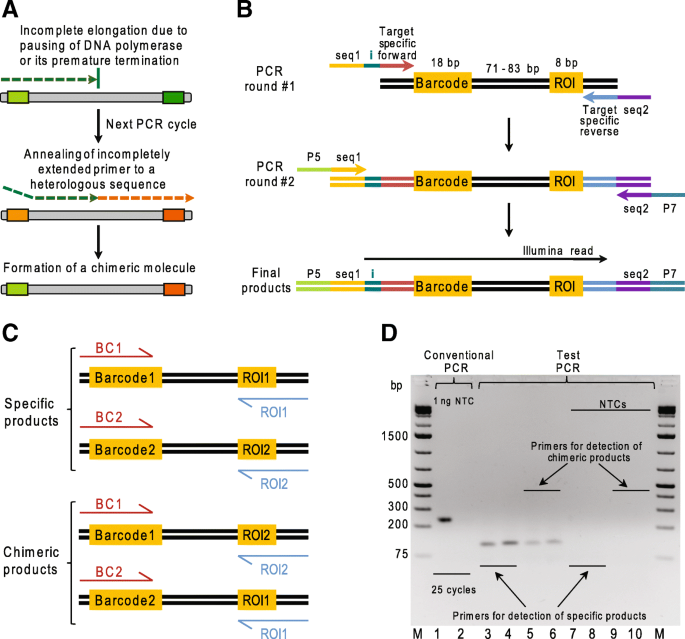
Optimized PCR conditions minimizing the formation of chimeric DNA molecules from MPRA plasmid libraries | BMC Genomics | Full Text
![PDF] Sources of erroneous sequences and artifact chimeric reads in next generation sequencing of genomic DNA from formalin-fixed paraffin-embedded samples | Semantic Scholar PDF] Sources of erroneous sequences and artifact chimeric reads in next generation sequencing of genomic DNA from formalin-fixed paraffin-embedded samples | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/3bcbbda66a2159f378b060ed33752d5d42657c72/5-Figure2-1.png)