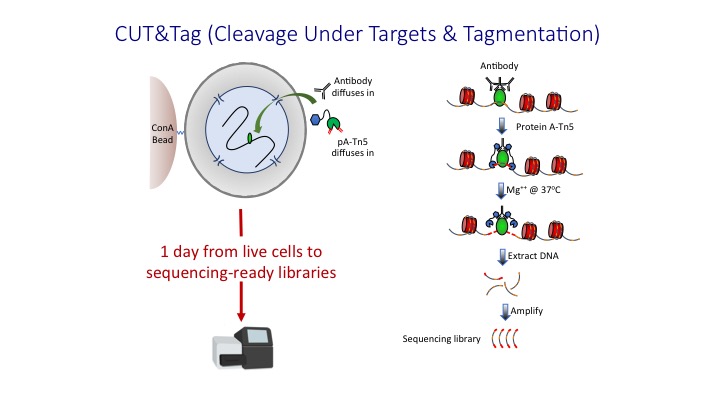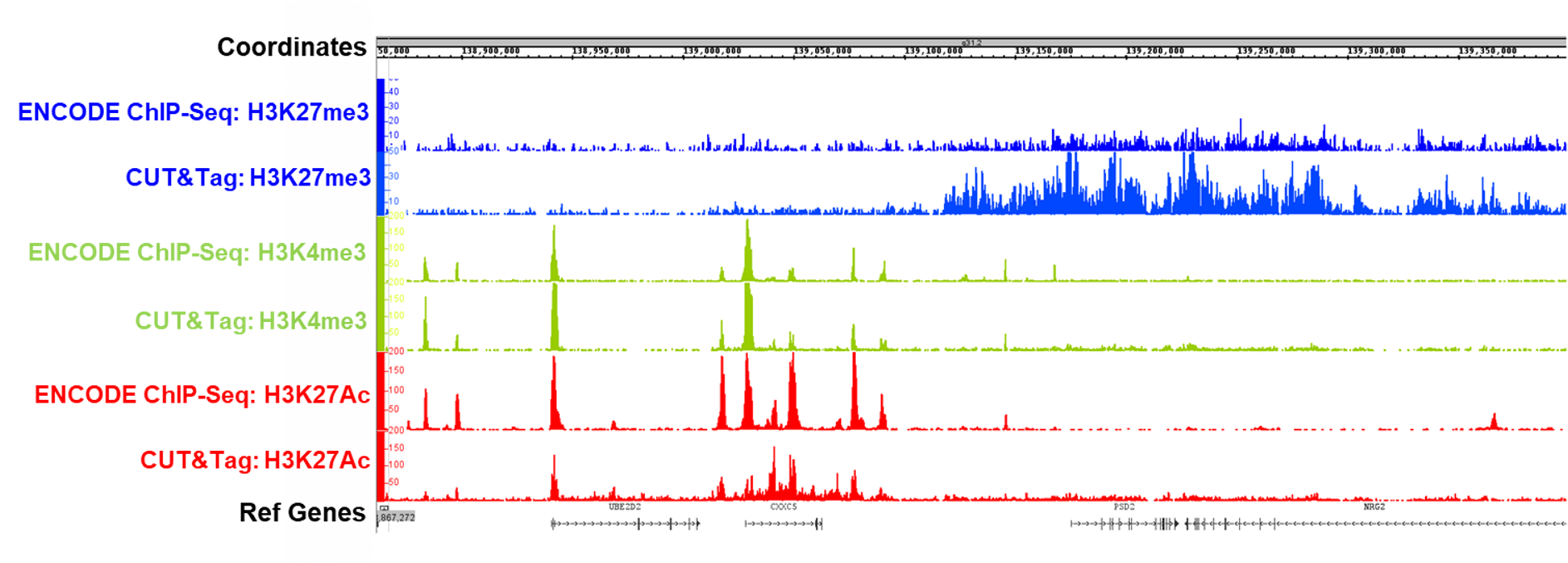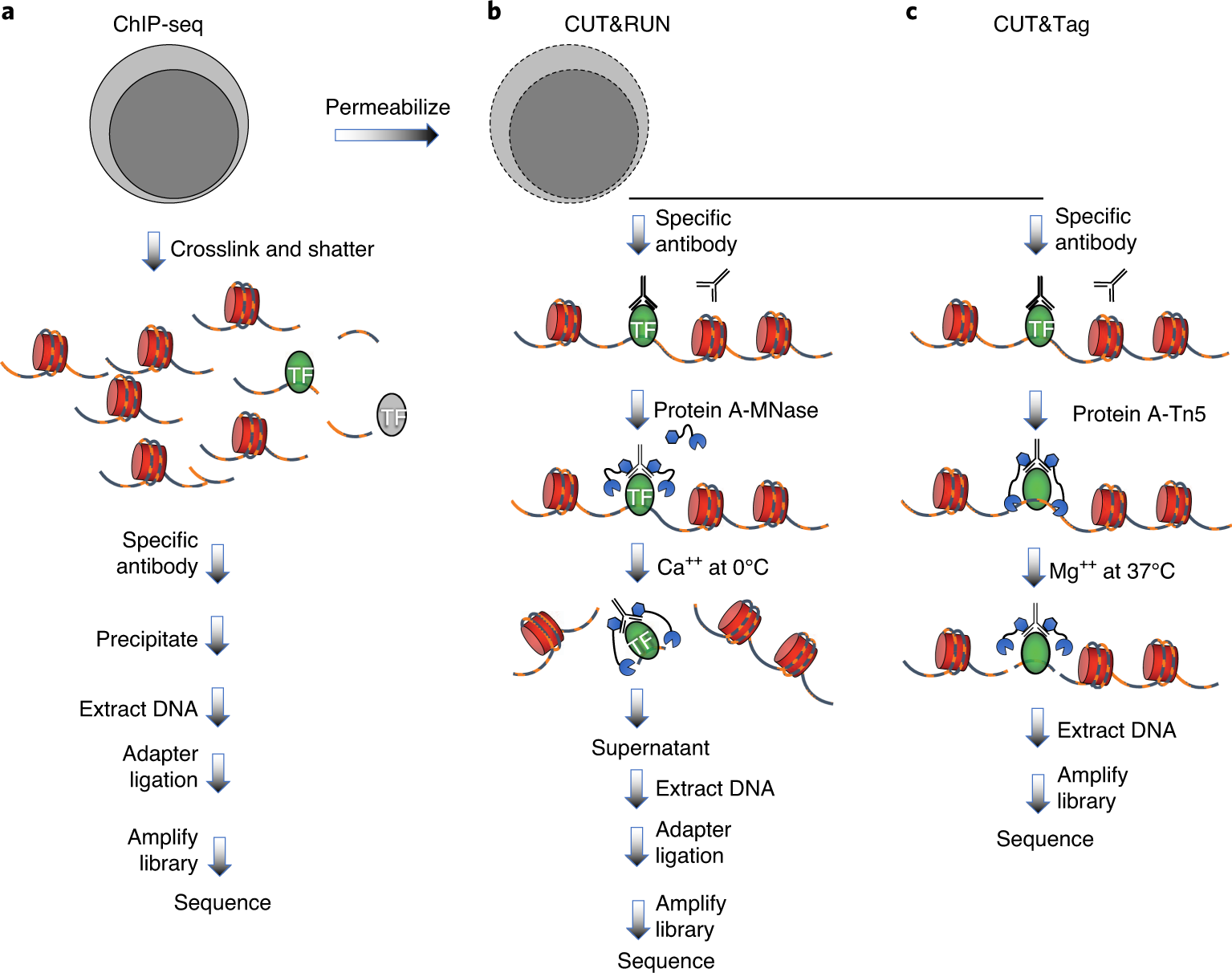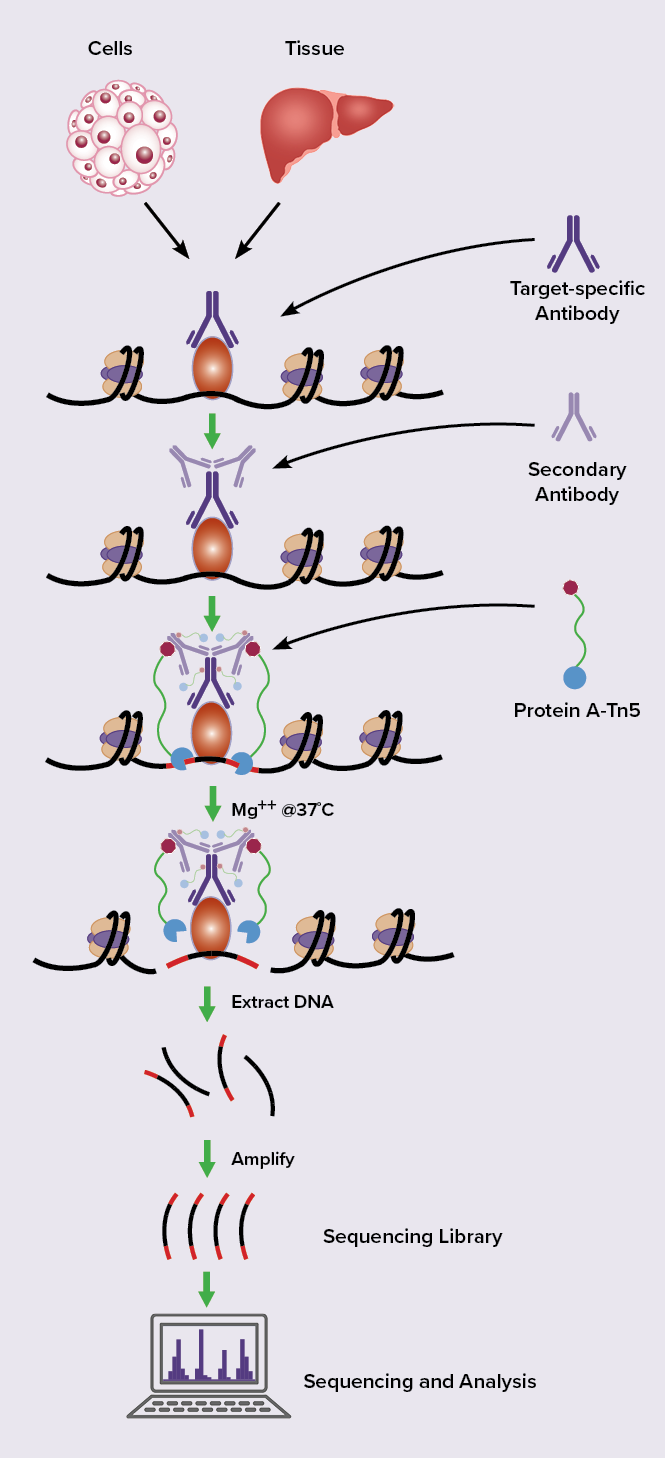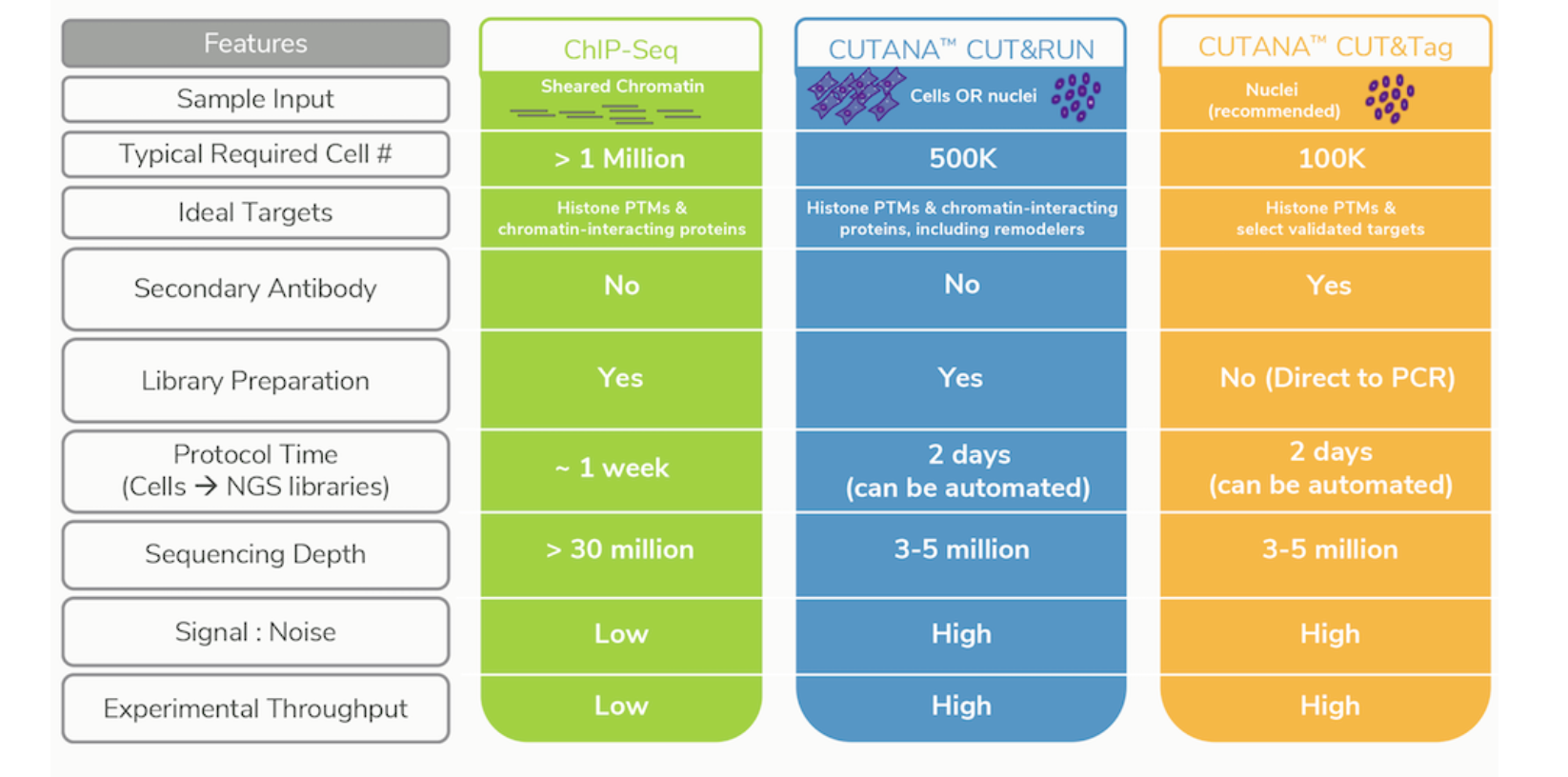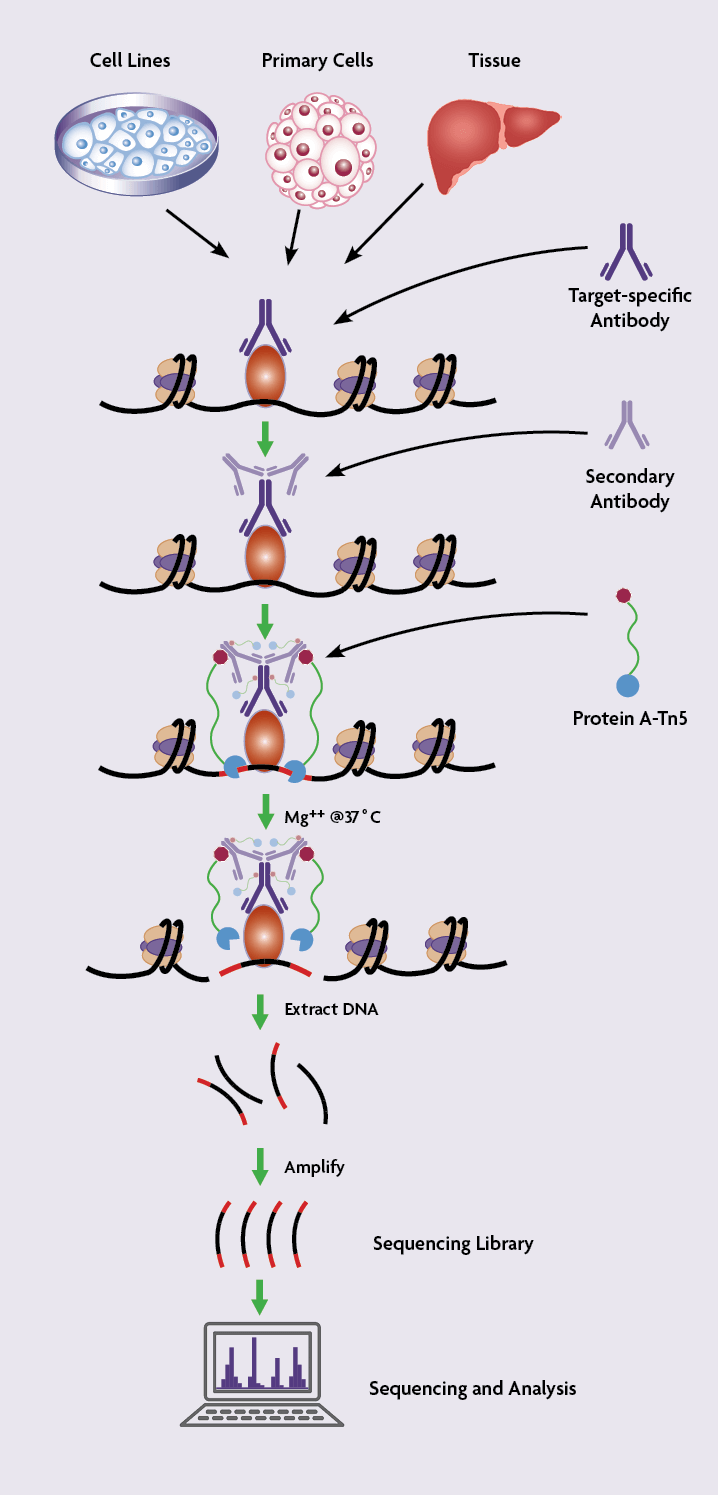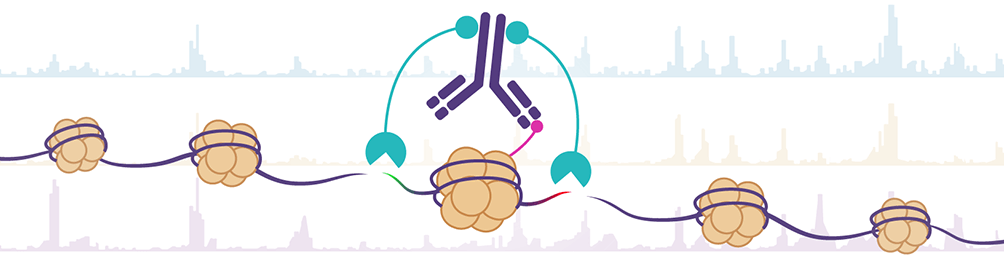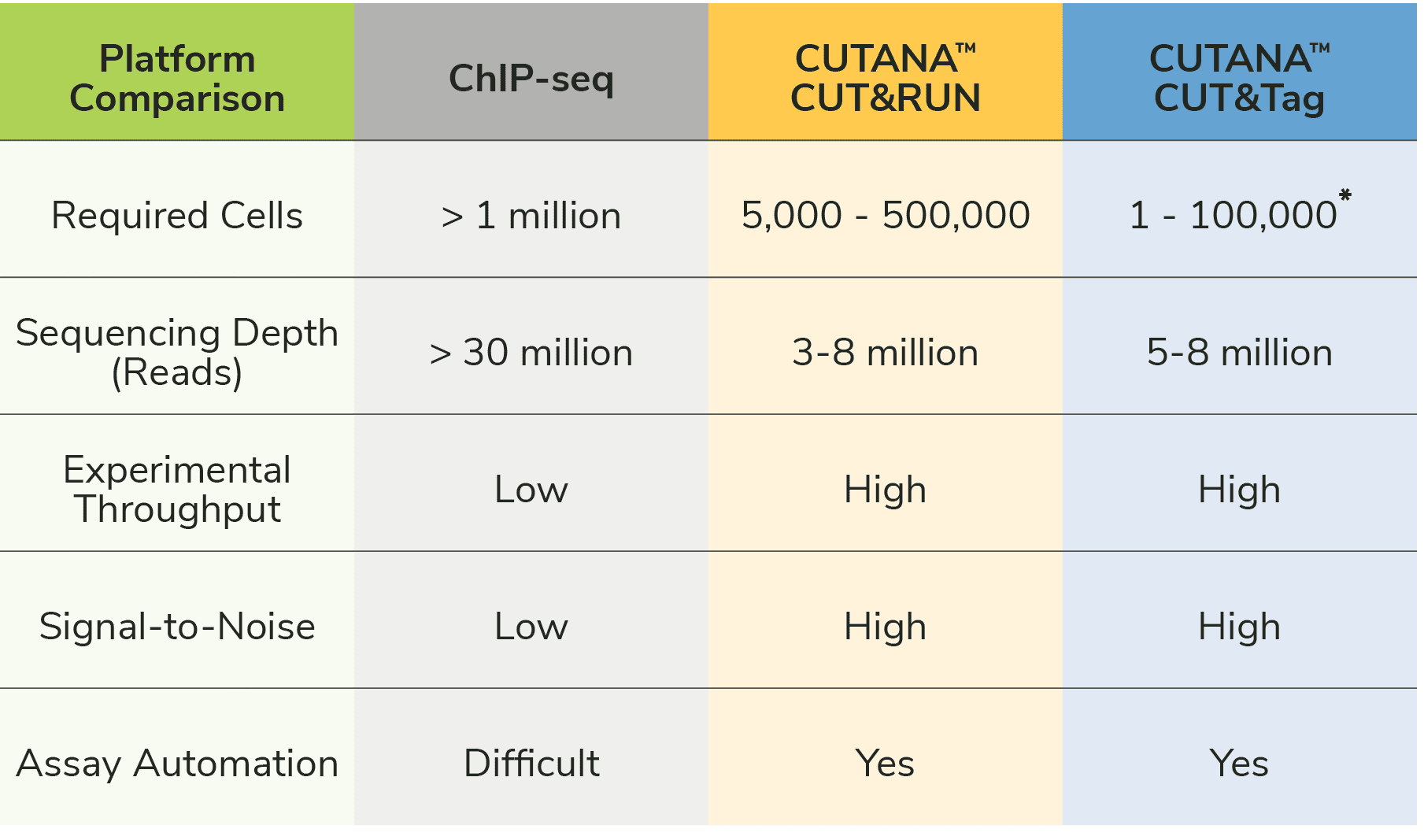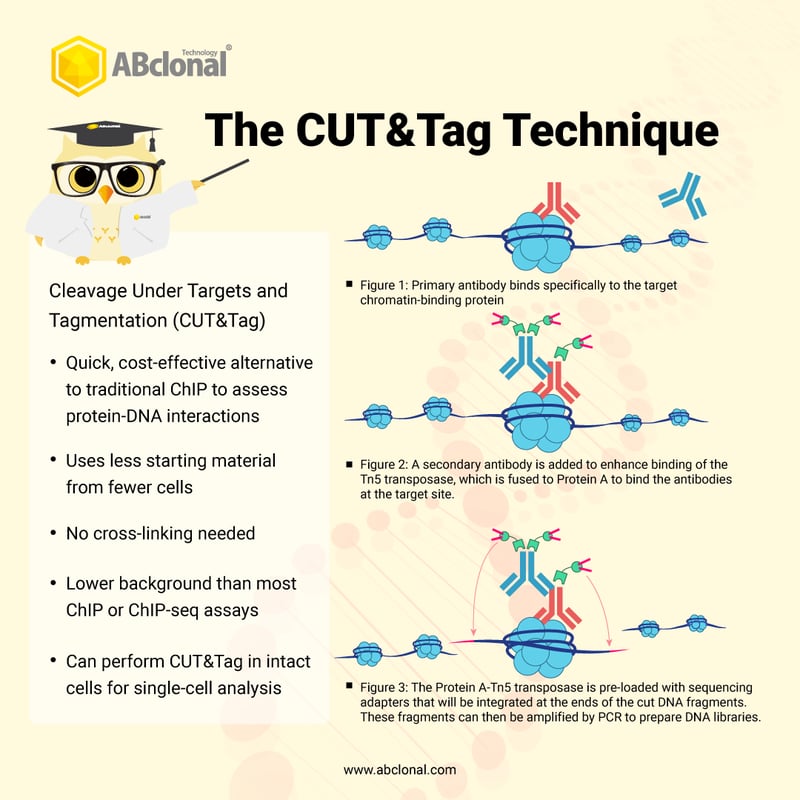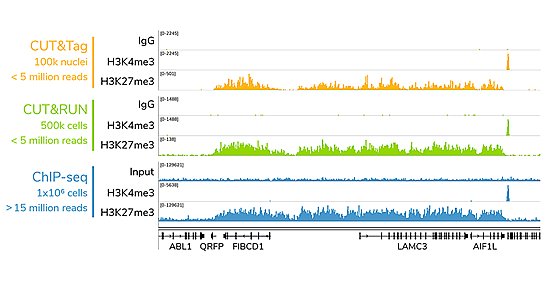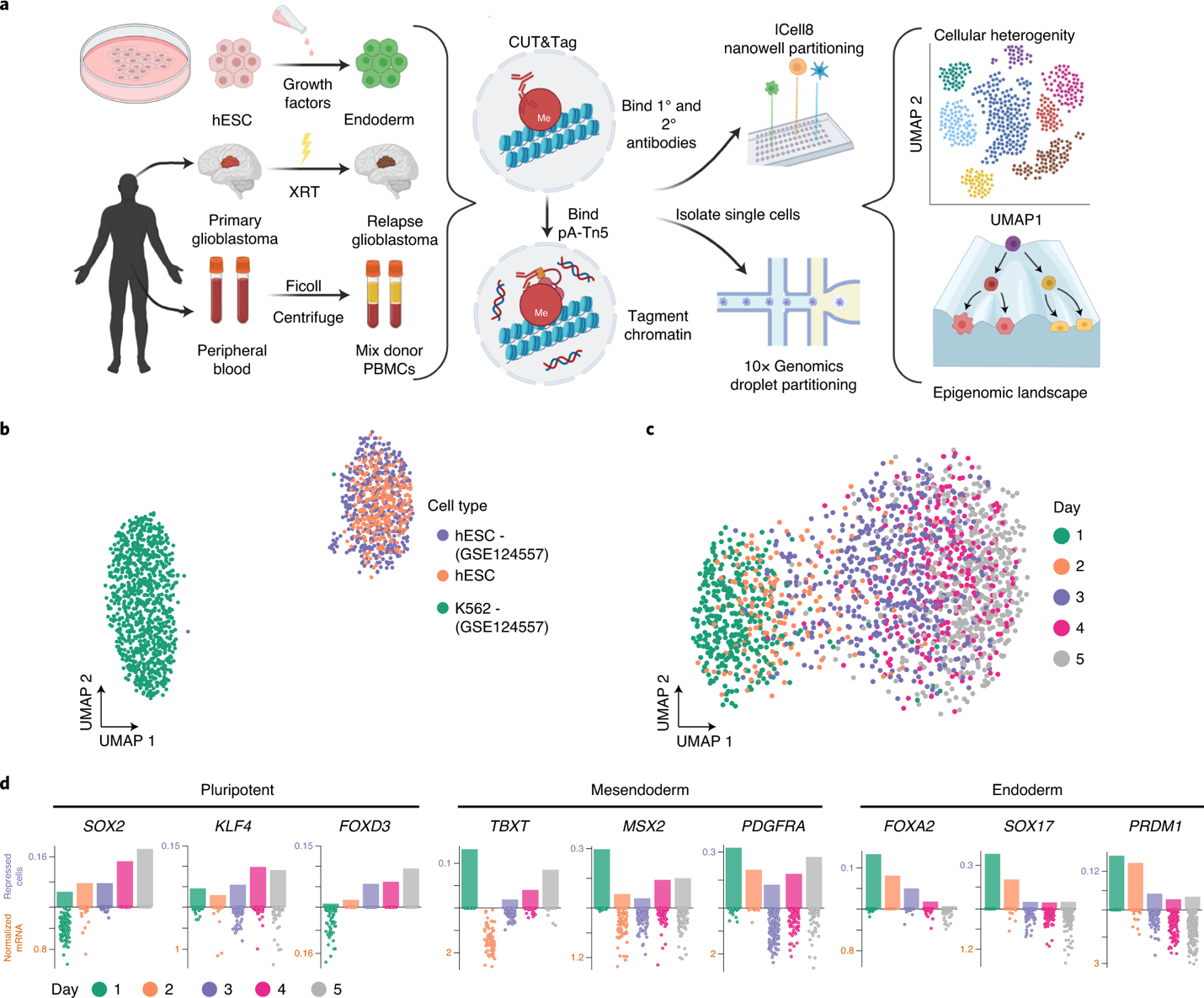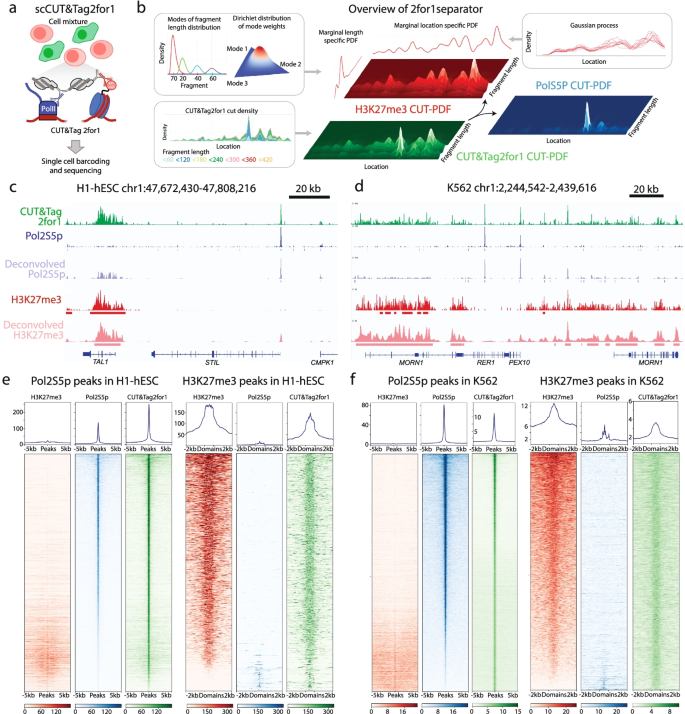
CUT&Tag2for1: a modified method for simultaneous profiling of the accessible and silenced regulome in single cells | Genome Biology | Full Text

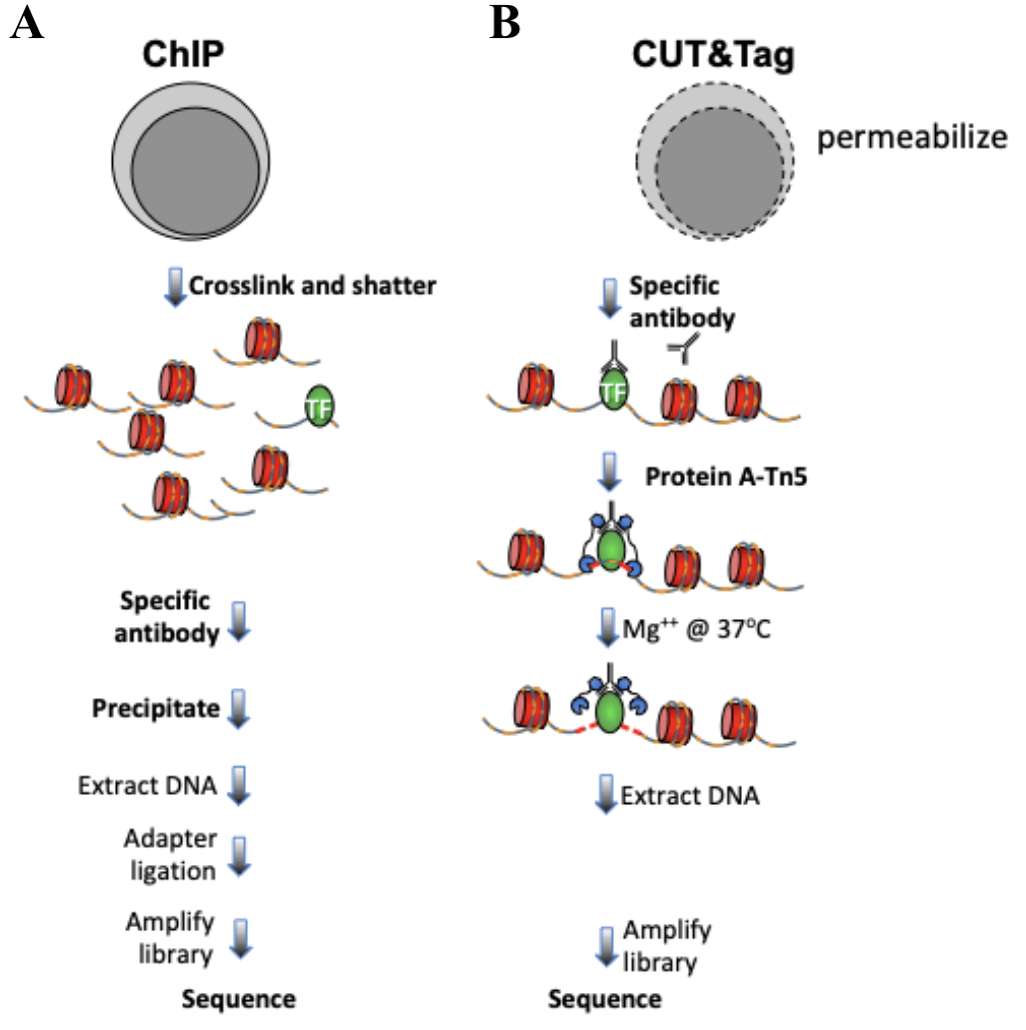
CUT&Tag for efficient epigenomic profiling of small samples and single cells | Nature Communications
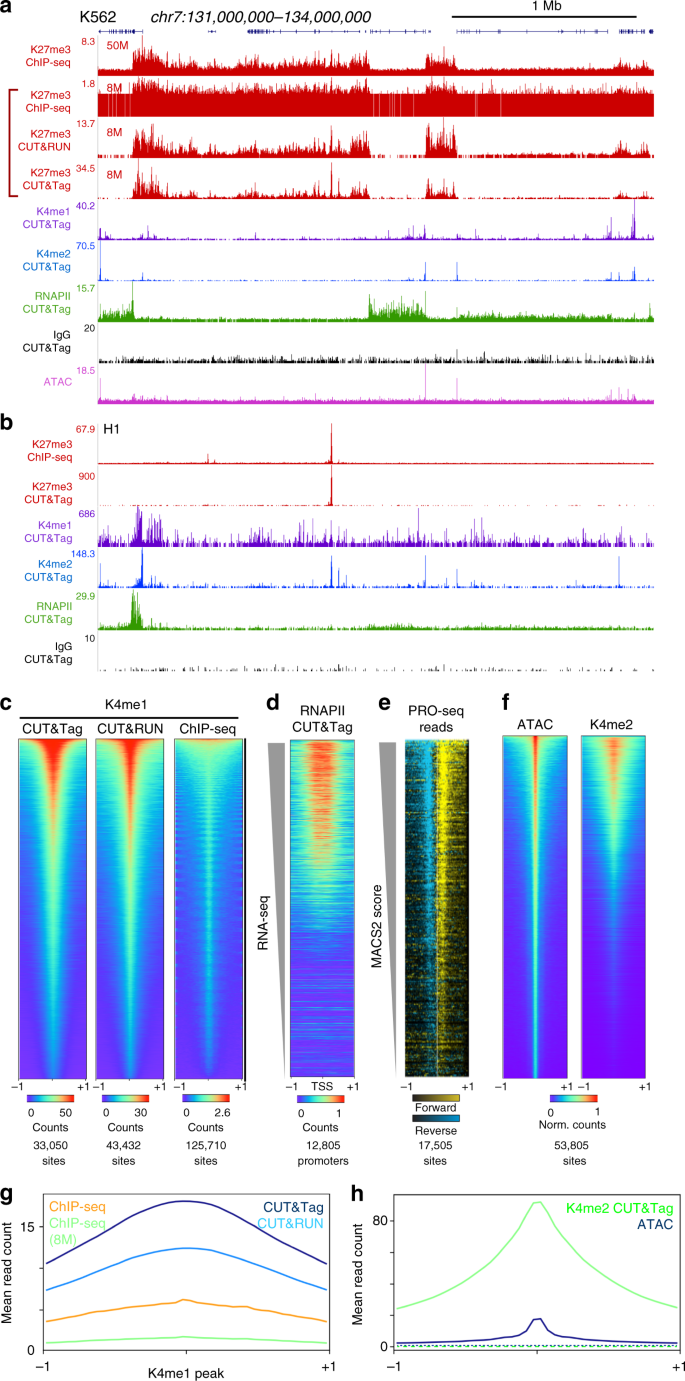
CUT&Tag for efficient epigenomic profiling of small samples and single cells | Nature Communications

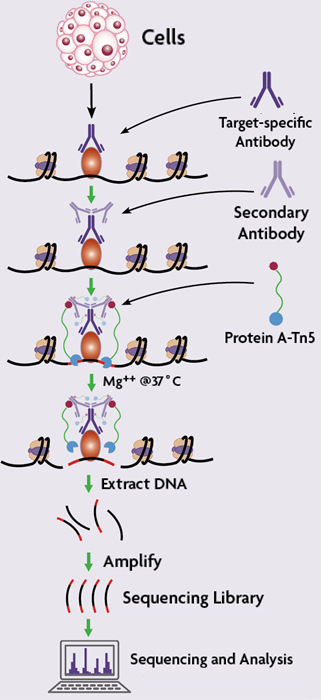
Overview of TIP-seq, a robust low-cell mapping method that combines... | Download Scientific Diagram

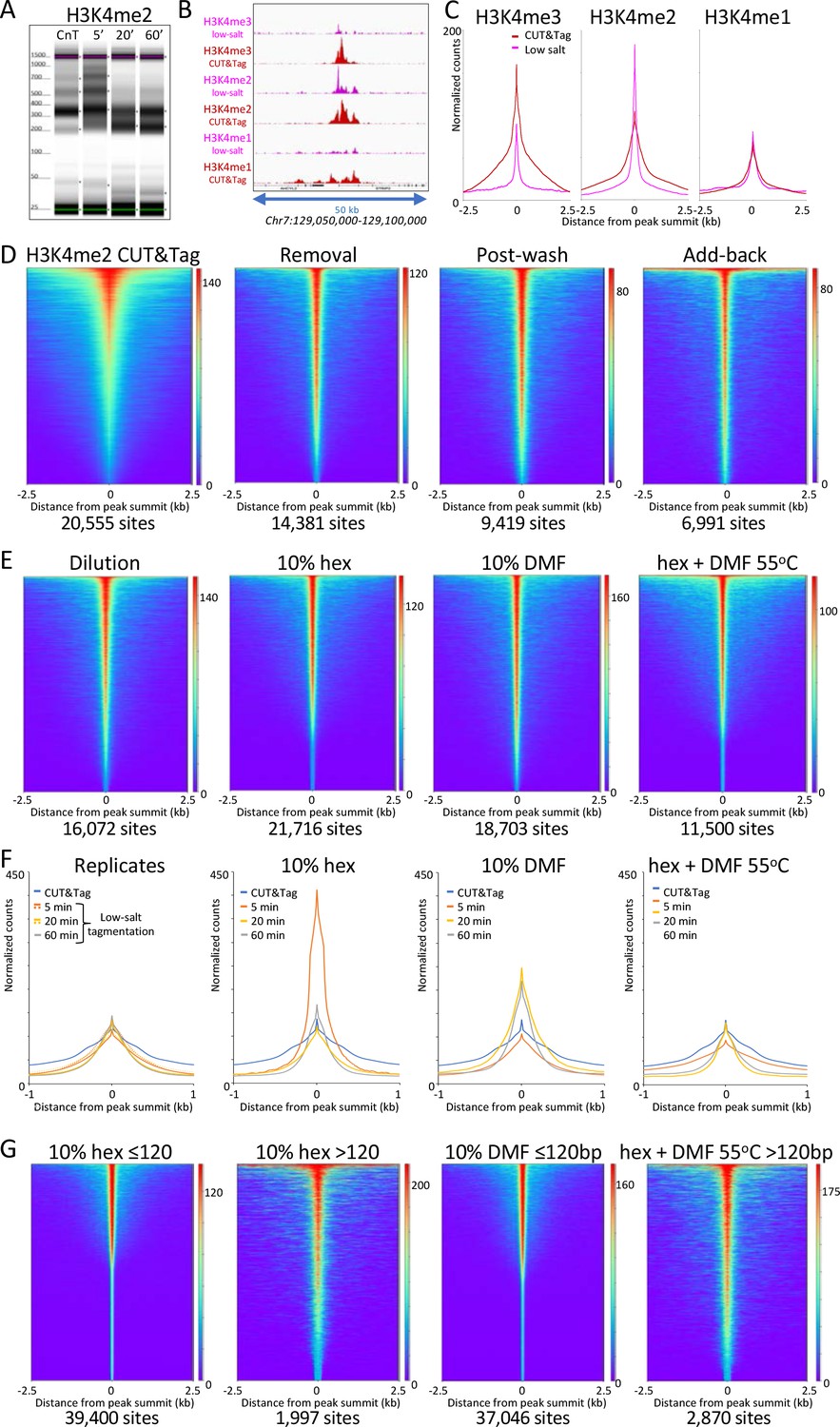
CUT&Tag with low-salt tagmentation (CUTAC). Steps in gray are lab-based... | Download Scientific Diagram

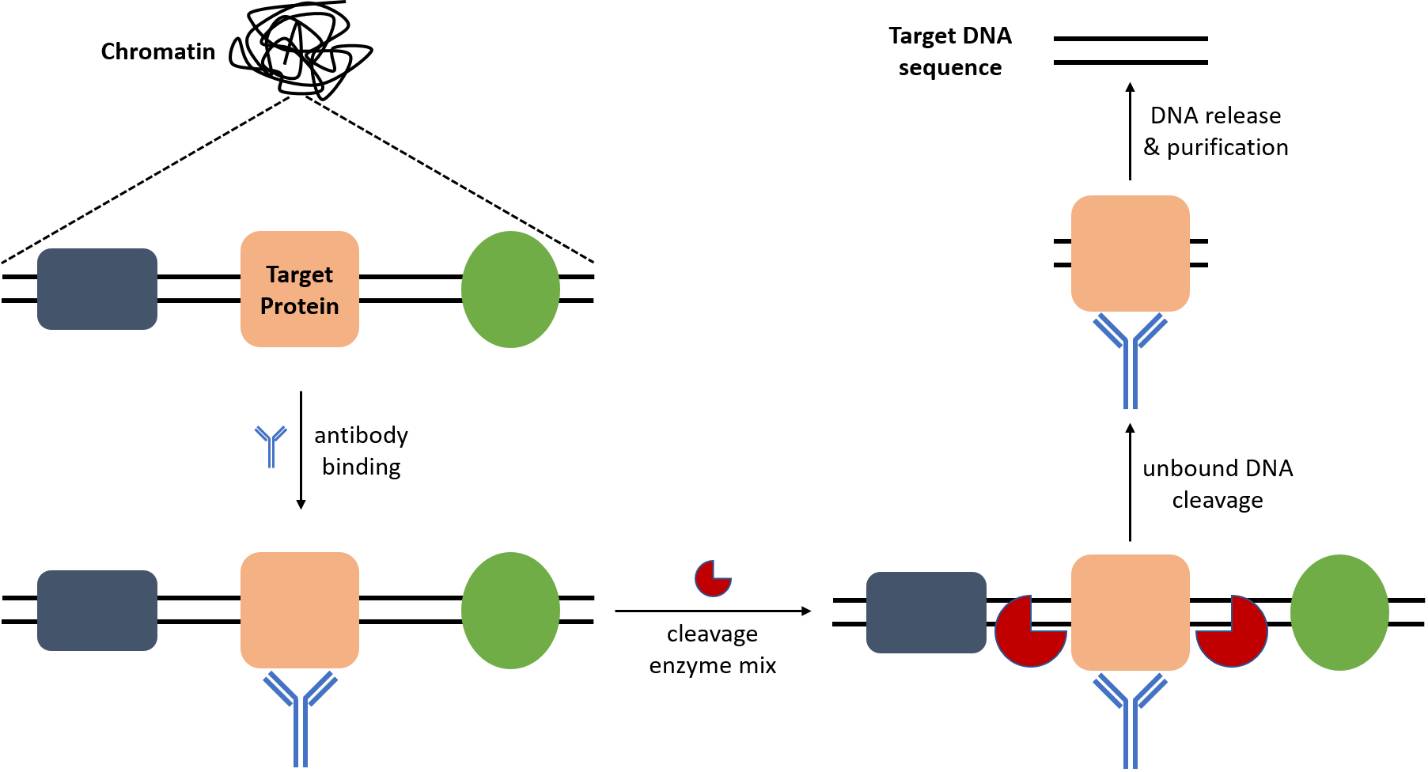
Mehdi Goudarzi on Twitter: "CUT&RUN method is great but is not well-suited for single-cell platforms. Hyperactive Tn5 transposase—Protein A (pA-Tn5) fusion protein loaded with sequencing adapters in the CUT&Tag protocol resolve this