NMD-degradome sequencing reveals ribosome-bound intermediates with 3′-end non-templated nucleotides | Nature Structural & Molecular Biology
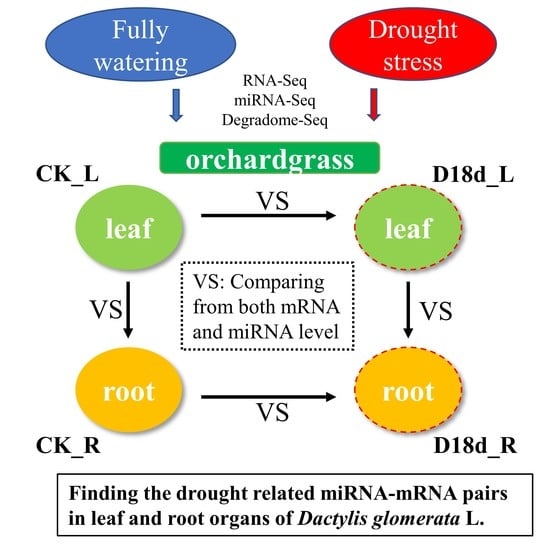
IJMS | Free Full-Text | Combinations of Small RNA, RNA, and Degradome Sequencing Uncovers the Expression Pattern of microRNA–mRNA Pairs Adapting to Drought Stress in Leaf and Root of Dactylis glomerata L.

miRNA and Degradome Sequencing Identify miRNAs and Their Target Genes Involved in the Browning Inhibition of Fresh-Cut Apples by Hydrogen Sulfide | Journal of Agricultural and Food Chemistry

Cells | Free Full-Text | Transcriptome and Degradome Profiling Reveals a Role of miR530 in the Circadian Regulation of Gene Expression in Kalanchoë marnieriana
Small RNA and Degradome Sequencing Reveal Complex Roles of miRNAs and Their Targets in Developing Wheat Grains | PLOS ONE

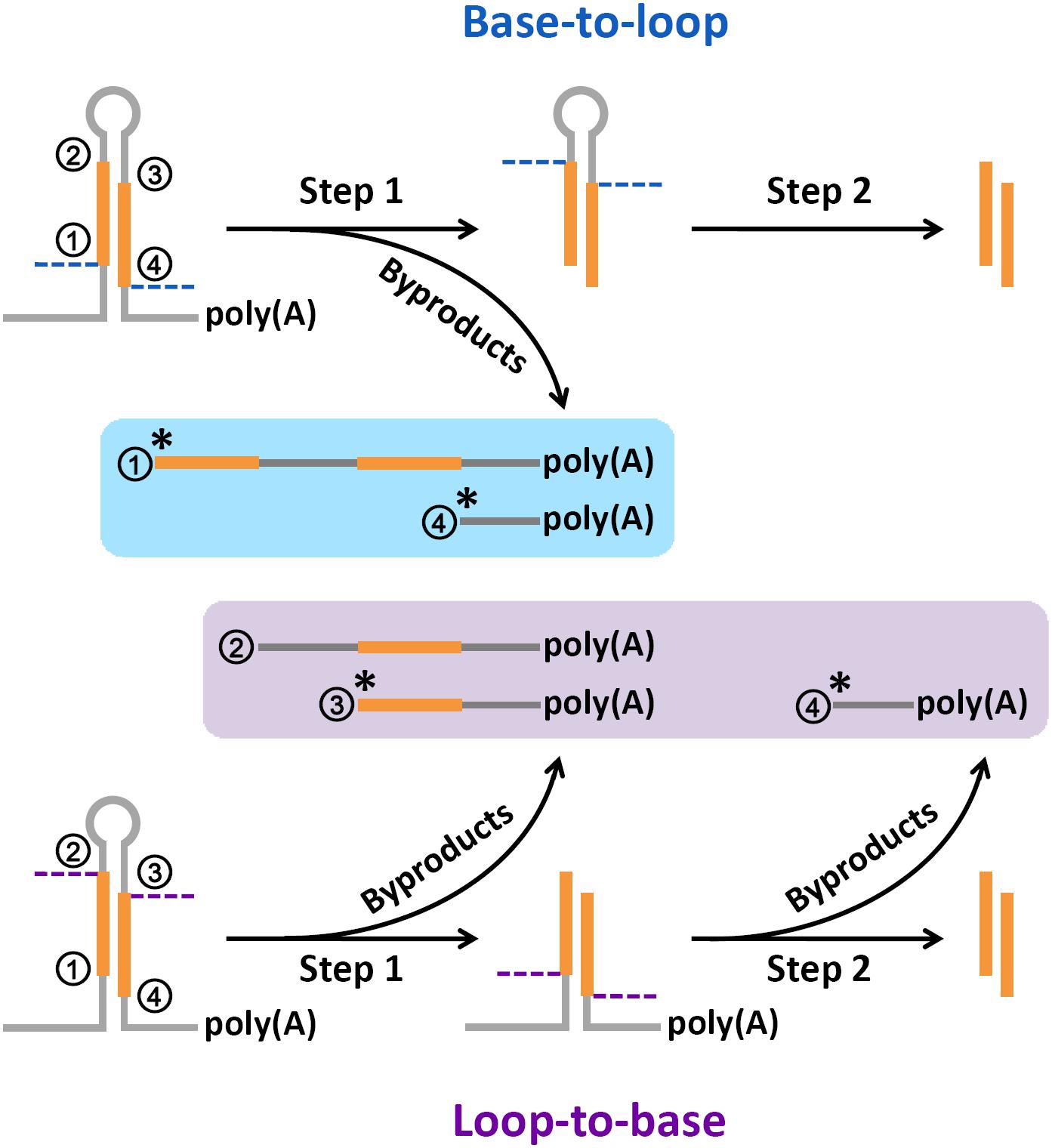
An improved method of constructing degradome library suitable for sequencing using Illumina platform | Plant Methods | Full Text

Comparative Small RNA and Degradome Sequencing Provides Insights into Antagonistic Interactions in the Biocontrol Fungus Clonostachys rosea | Applied and Environmental Microbiology
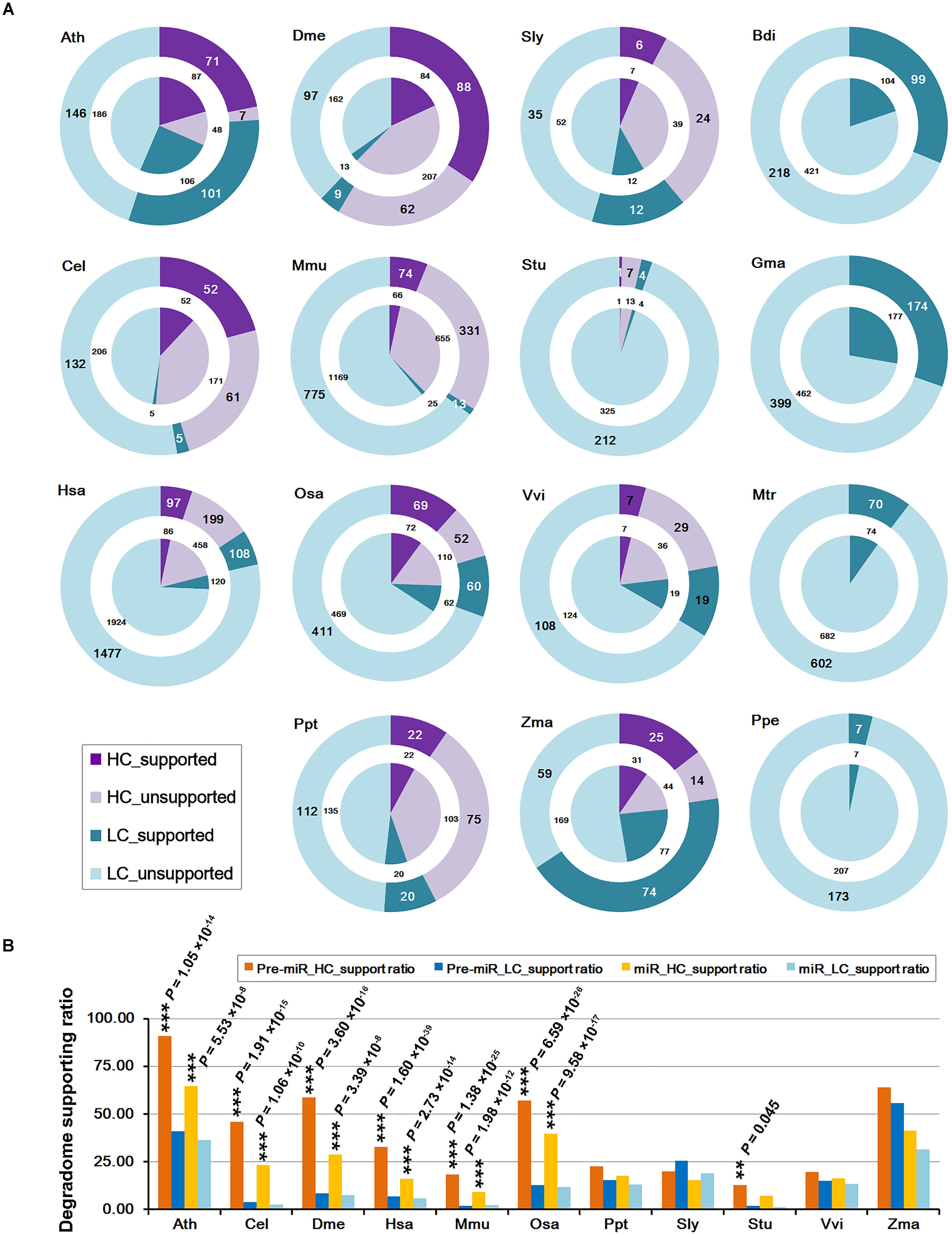
Degradome sequencing validated that three known miRNAs and one novel... | Download Scientific Diagram
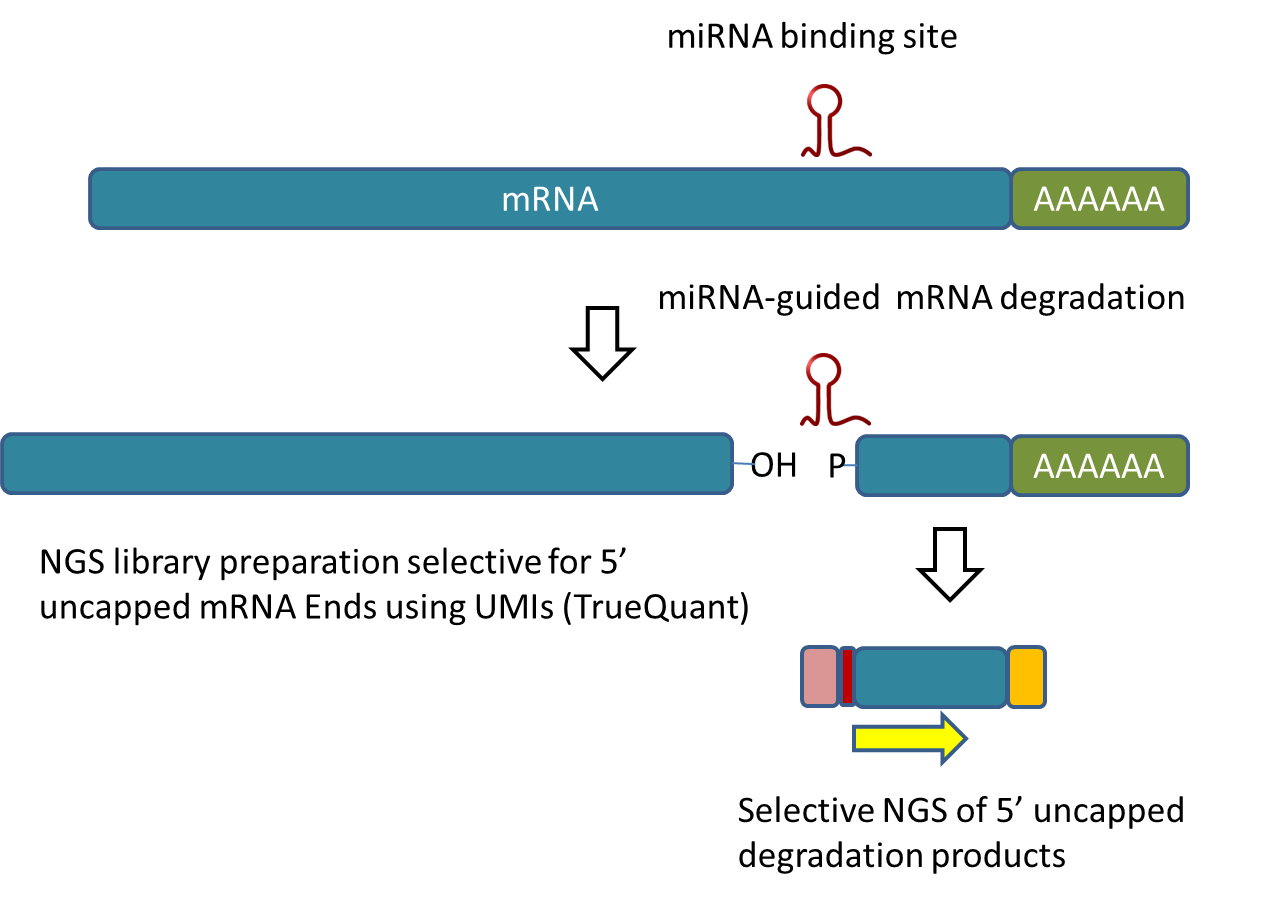
Fast and Efficient 5'P Degradome Library Preparation for Analysis of Co-Translational Decay in Arabidopsis
Small RNA and Degradome Sequencing Reveal Complex Roles of miRNAs and Their Targets in Developing Wheat Grains | PLOS ONE

High-throughput 5′P sequencing enables the study of degradation-associated ribosome stalls - ScienceDirect

Transcriptional multiomics reveals the mechanism of seed deterioration in Nicotiana tabacum L. and Oryza sativa L. - ScienceDirect

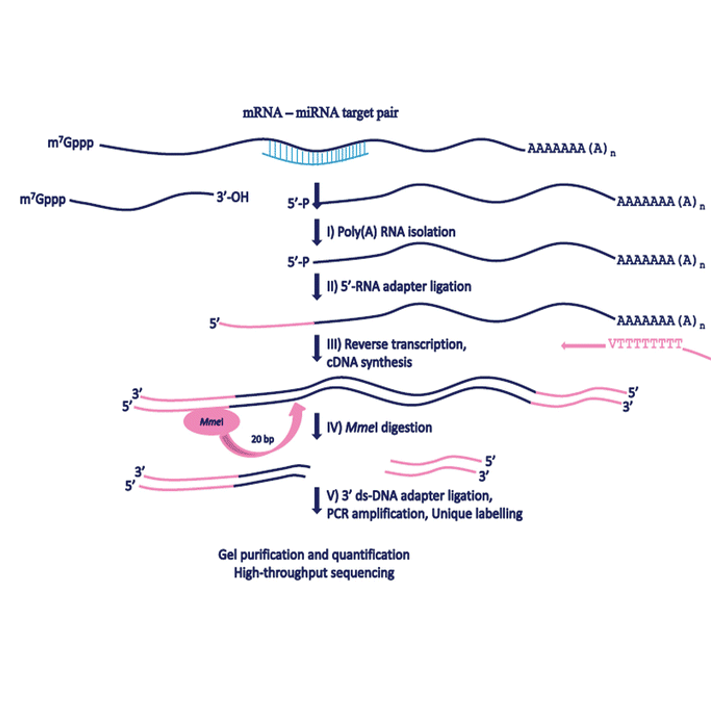
Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome | Nature Protocols