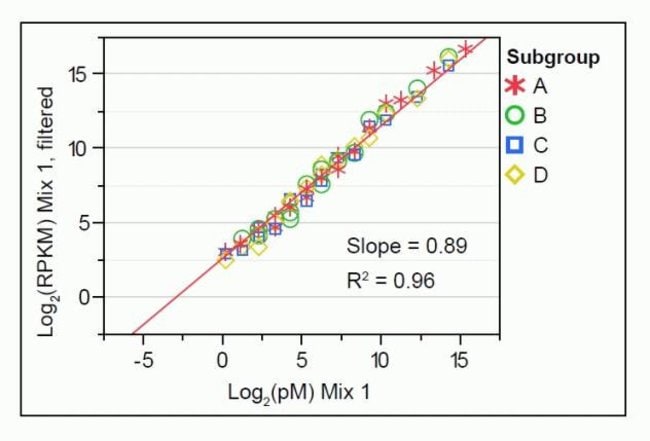
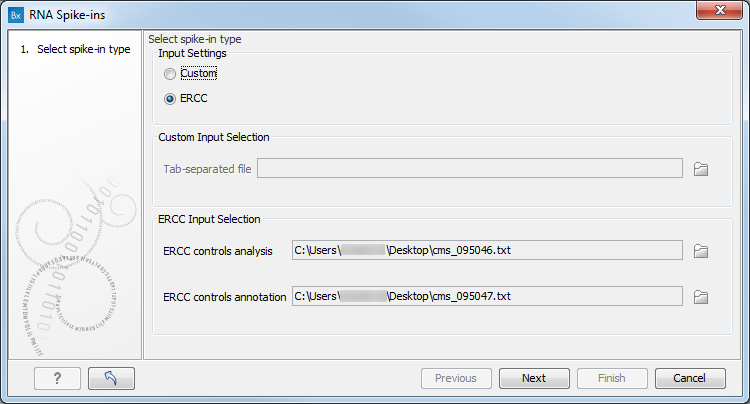
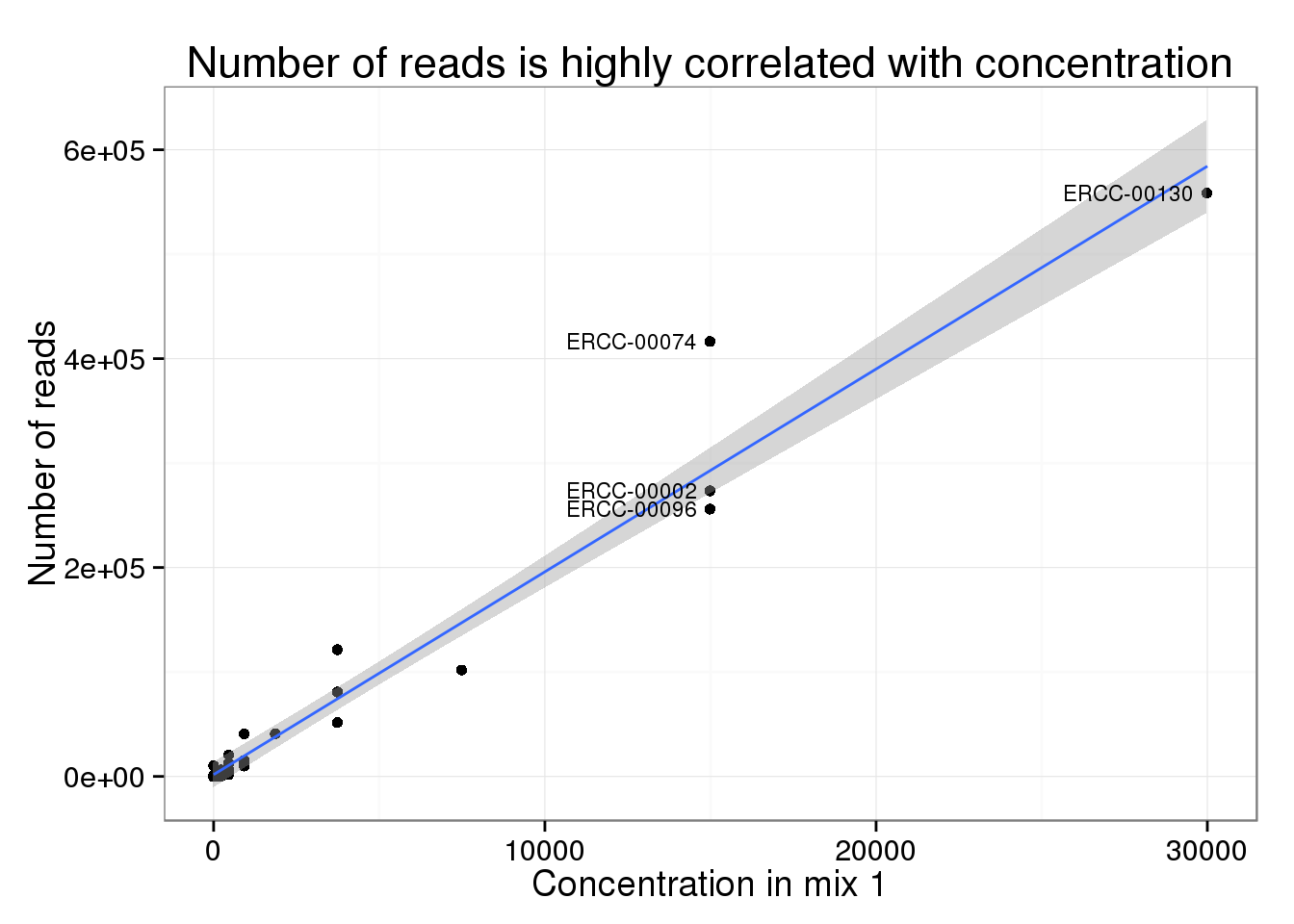
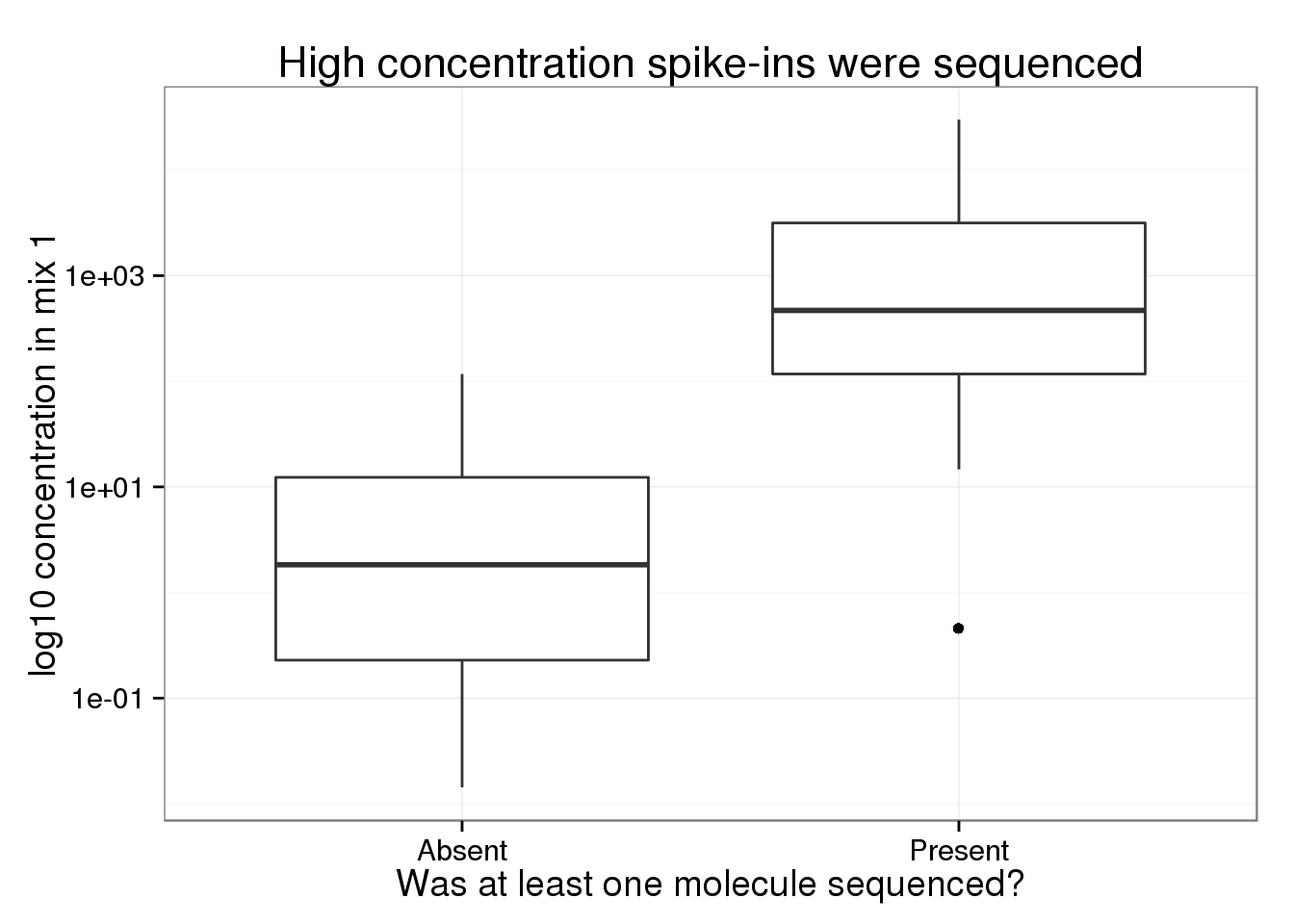
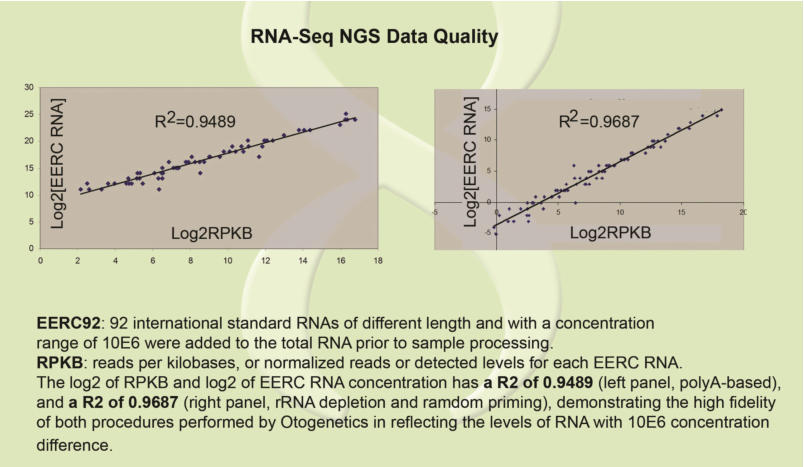
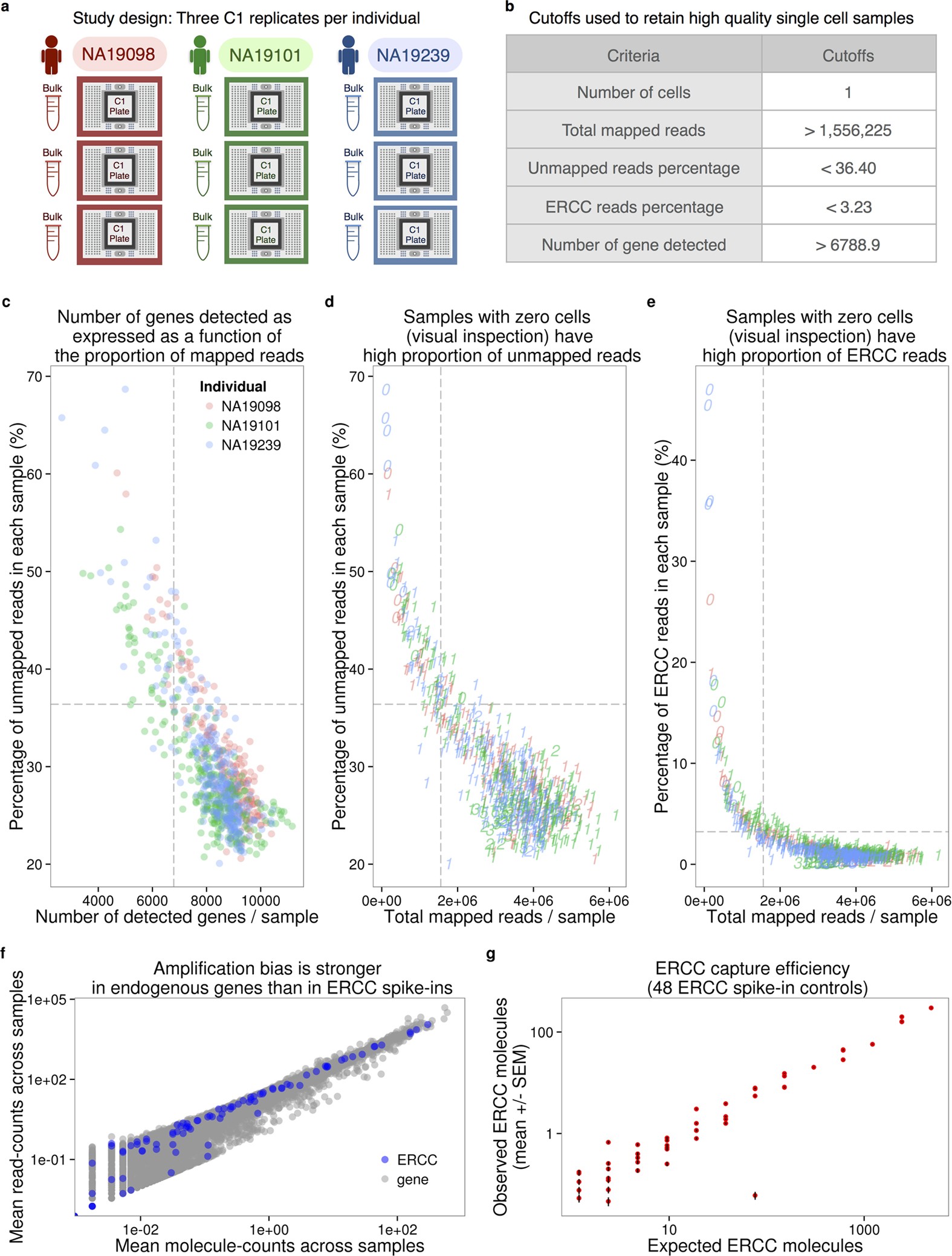
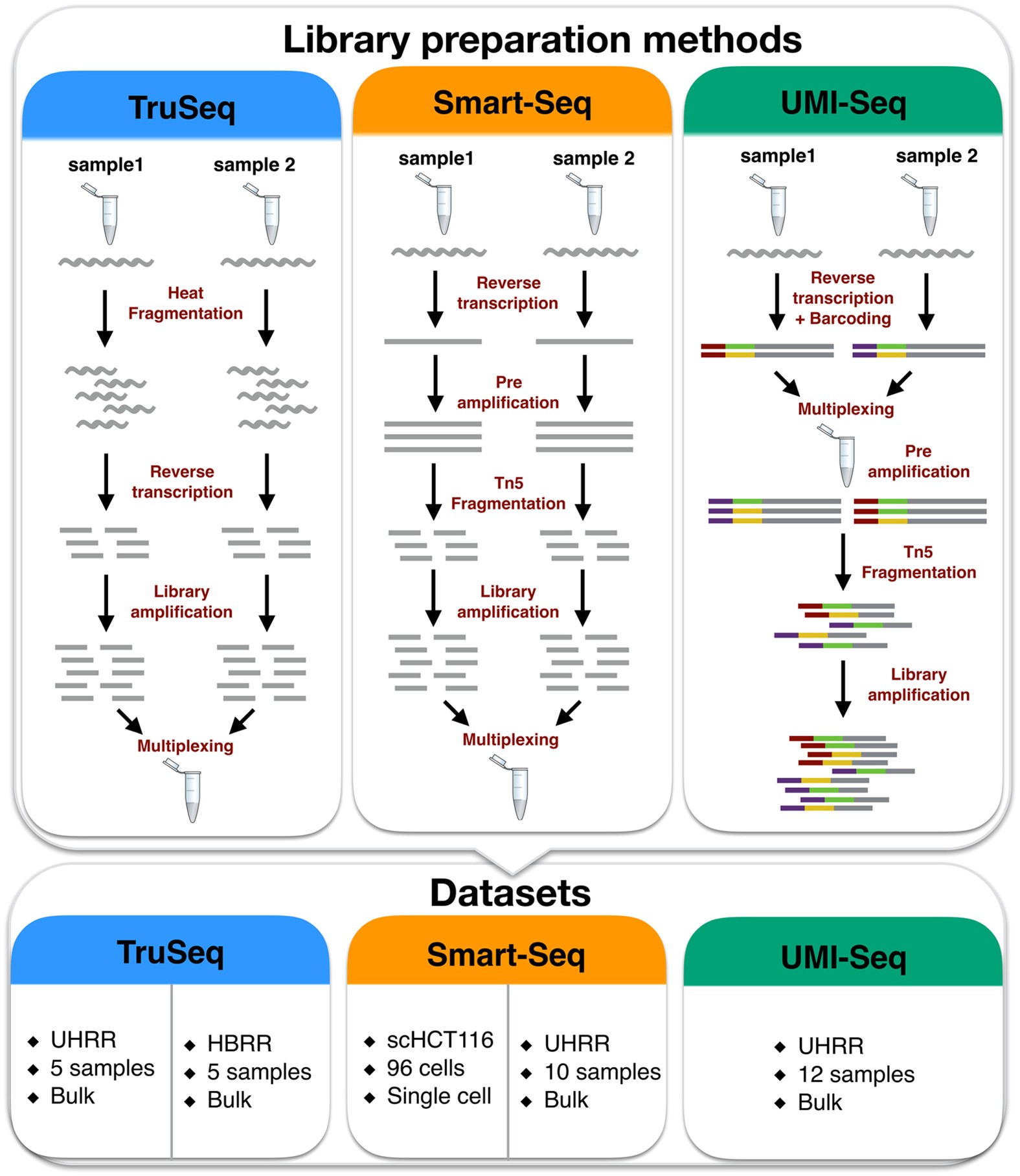
A complete statistical model for calibration of RNA-seq counts using external spike-ins and maximum likelihood theory | PLOS Computational Biology

Using the ERCC spike-in controls for normalization, zebrafish data set.... | Download Scientific Diagram

Lexogen has developed Spike-In RNA Variants (SIRVs) for the quantification of mRNA isoforms and begun a test program | RNA-Seq Blog

Analysis of ERCC spike-ins. Differential expression of ERCC spike-ins.... | Download Scientific Diagram
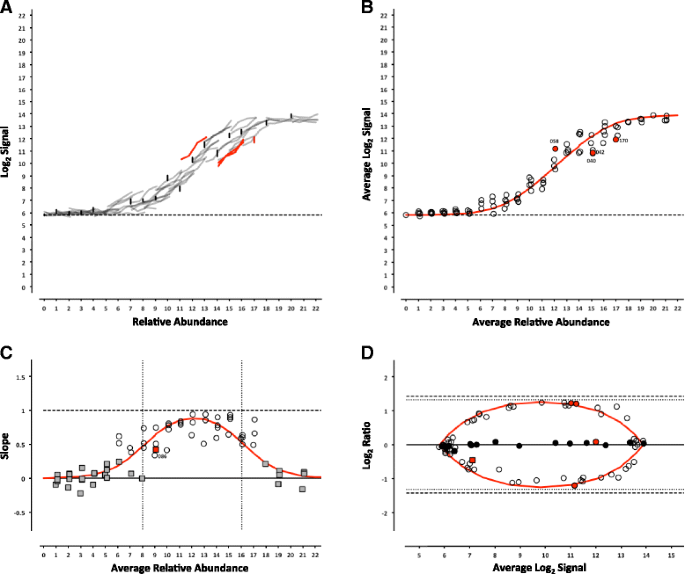
Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design | BMC Biotechnology | Full Text

Differential expression analysis of ERCC spike-ins. a Violin-plots of... | Download Scientific Diagram
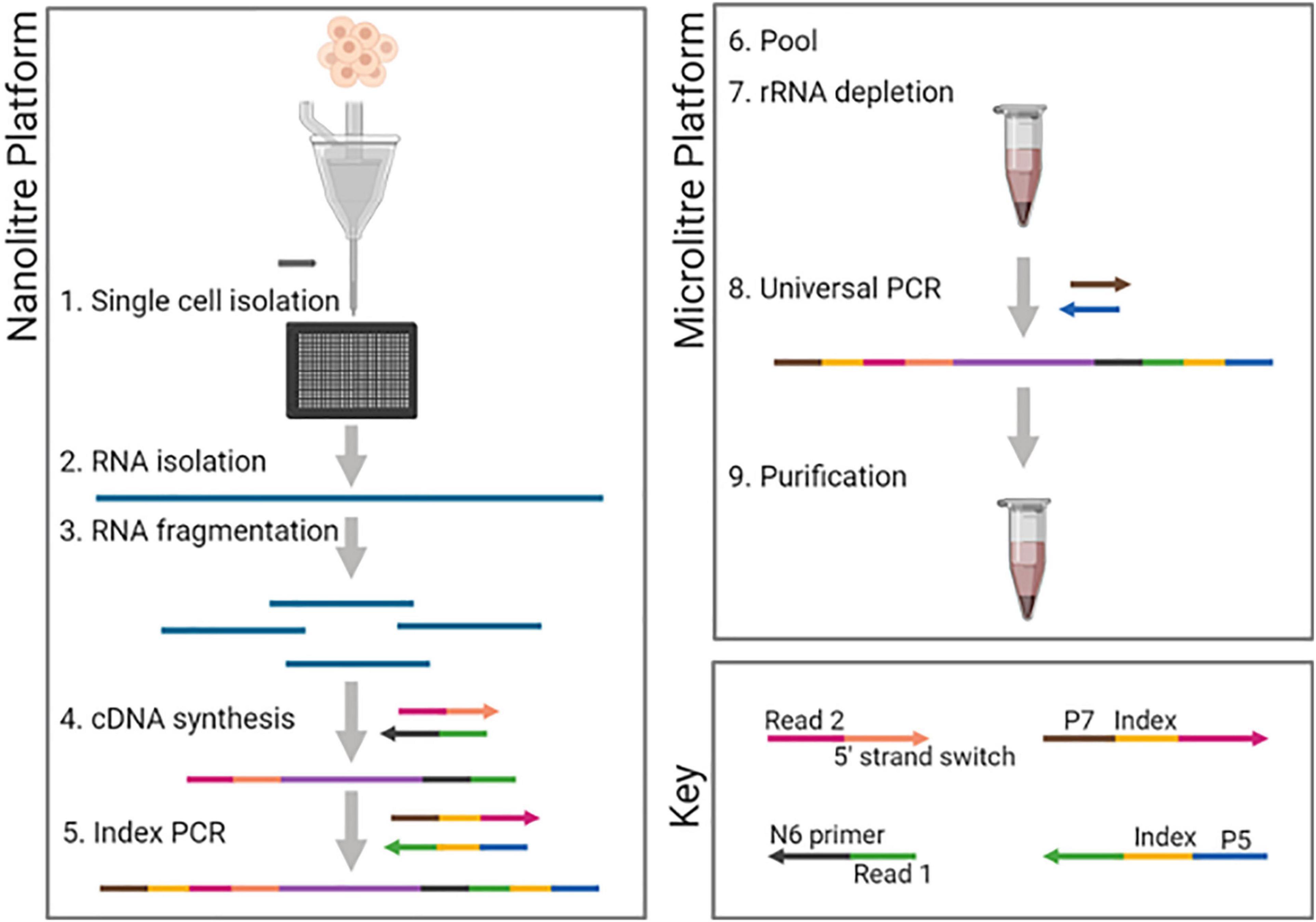
Frontiers | A Scalable Strand-Specific Protocol Enabling Full-Length Total RNA Sequencing From Single Cells
ERCC Spike-In Control Transcripts Provides Confidence in Agilent Microarray and RNA-Seq Gene Expression Data
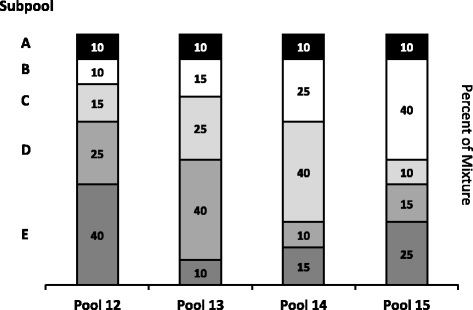
Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design | BMC Biotechnology | Full Text
ERCC Spike-In Control Transcripts Provides Confidence in Agilent Microarray and RNA-Seq Gene Expression Data

Lexogen has developed Spike-In RNA Variants (SIRVs) for the quantification of mRNA isoforms and begun a test program | RNA-Seq Blog