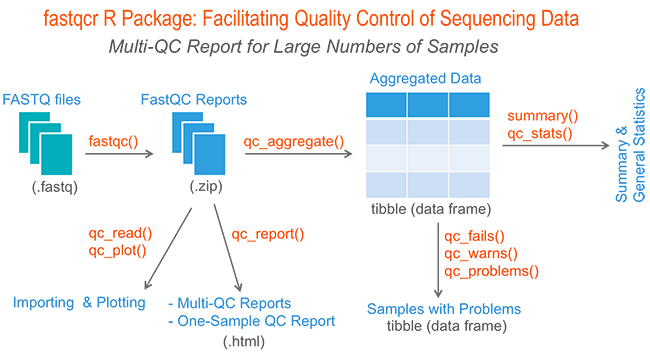
fastqcr: An R Package Facilitating Quality Controls of Sequencing Data for Large Numbers of Samples | R-bloggers

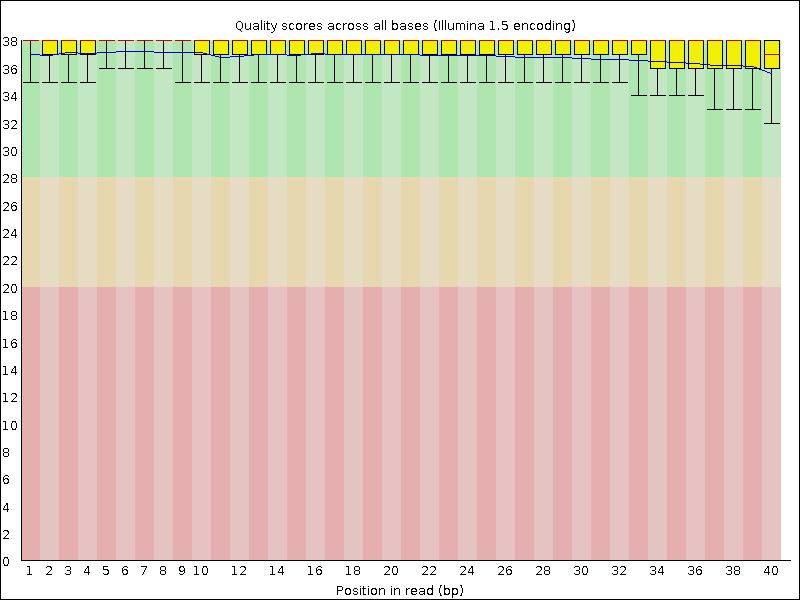
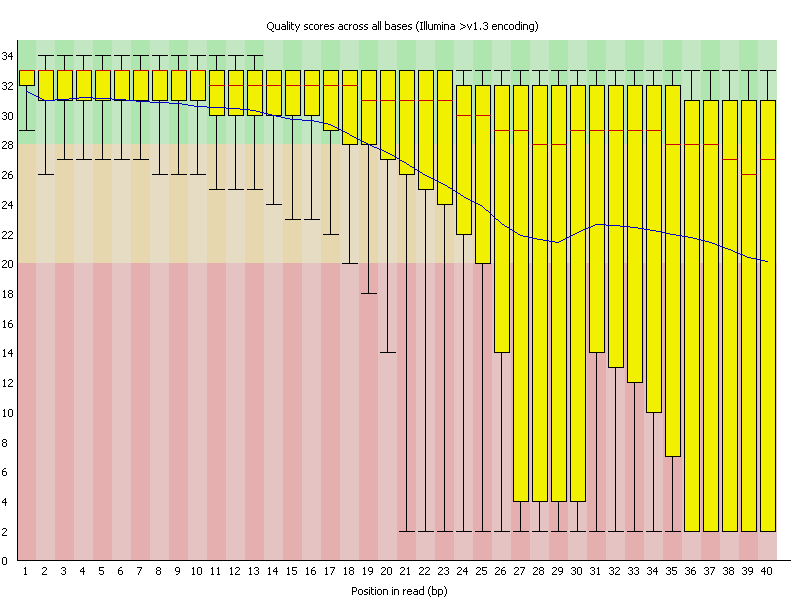
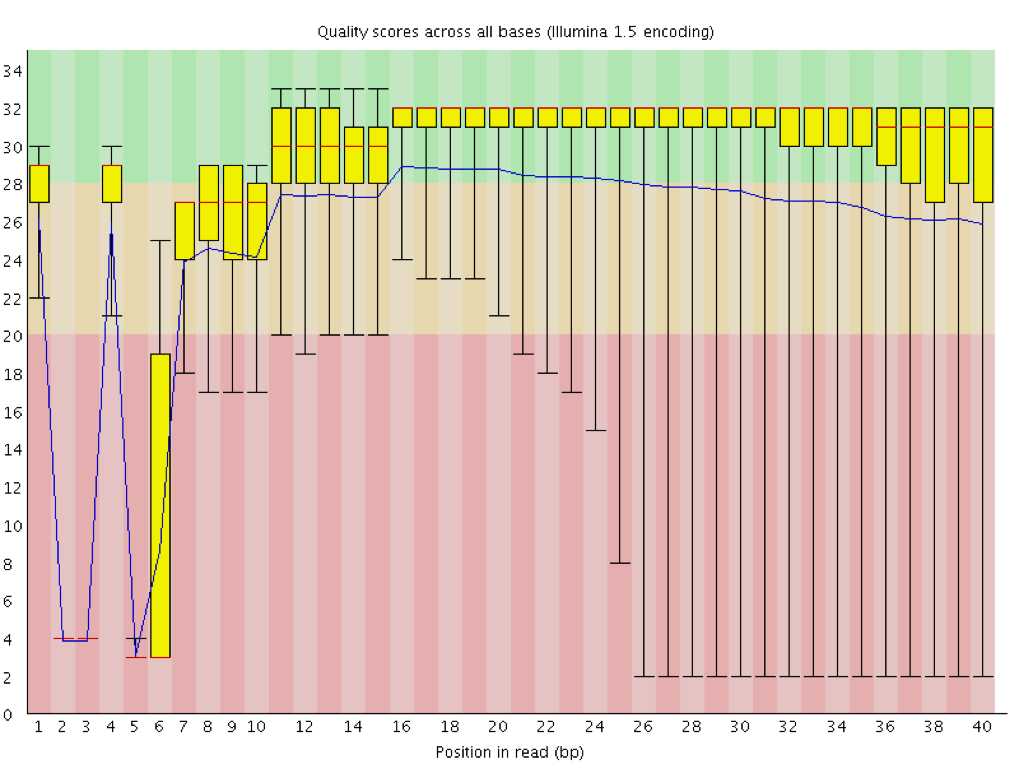
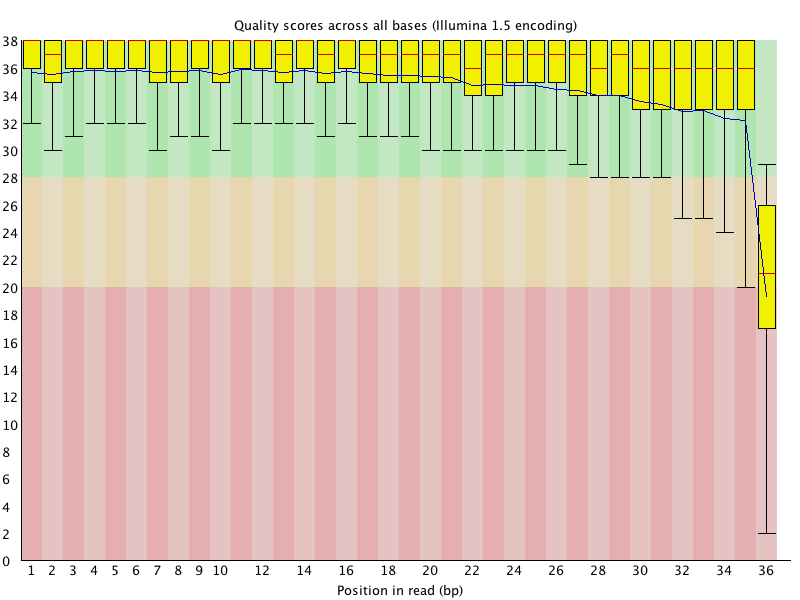
Box and whisker plot of the per base sequence quality generated by the... | Download Scientific Diagram

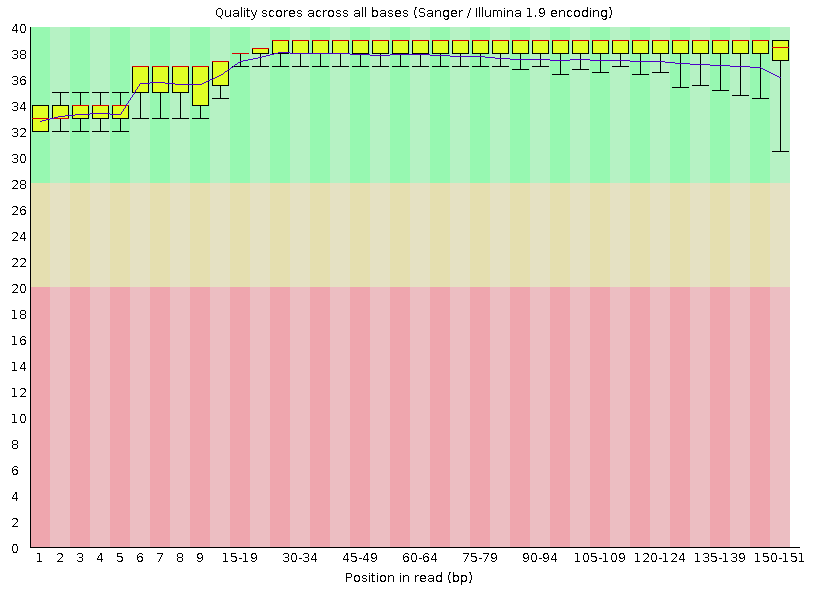
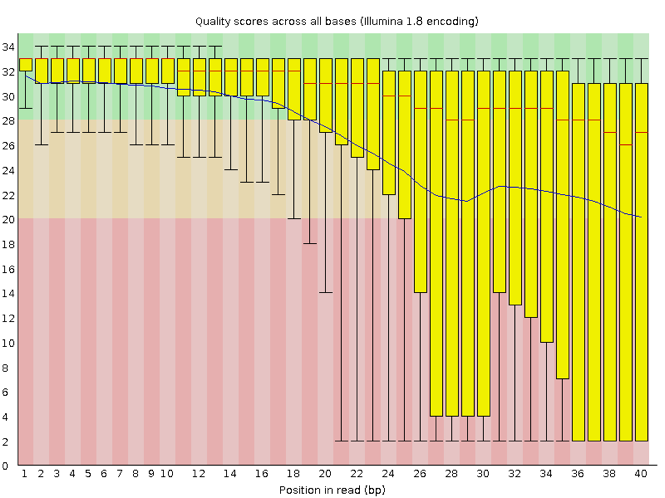
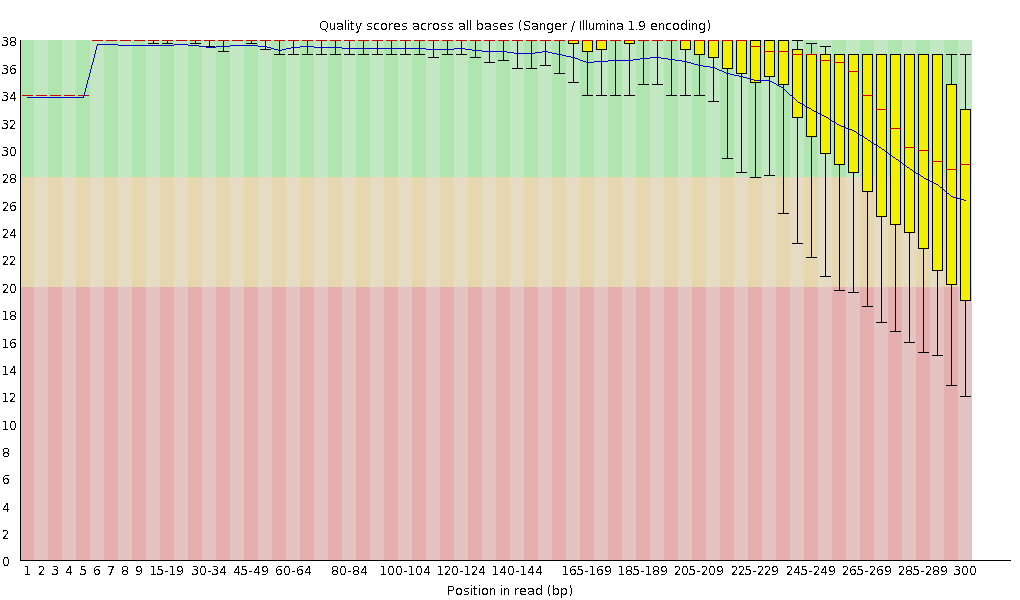
A representative example of per base sequence quality for RNA-Seq data... | Download Scientific Diagram
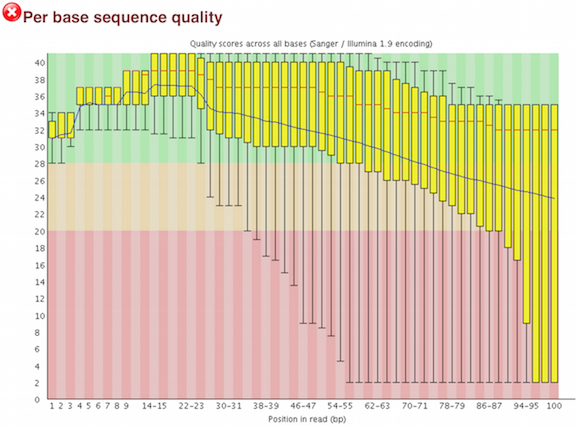
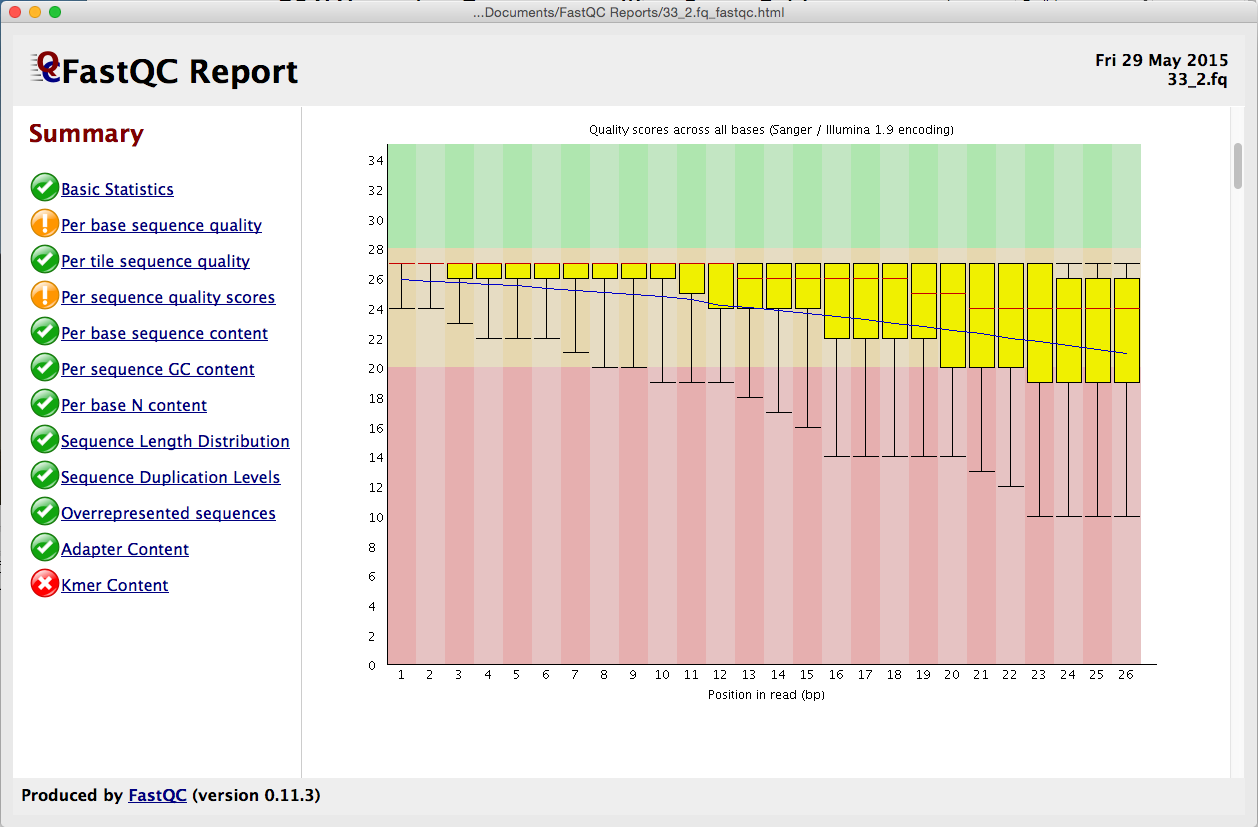
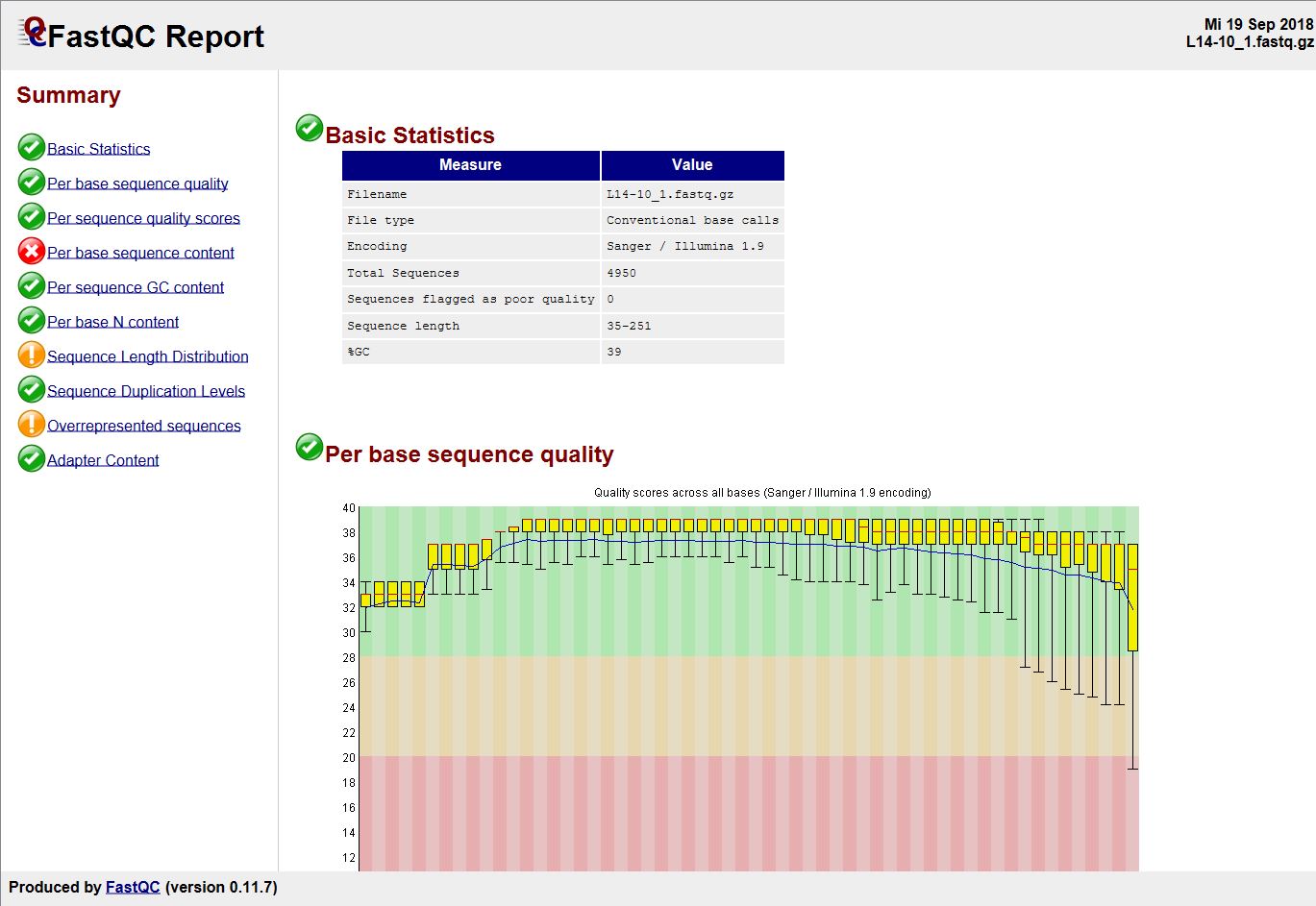
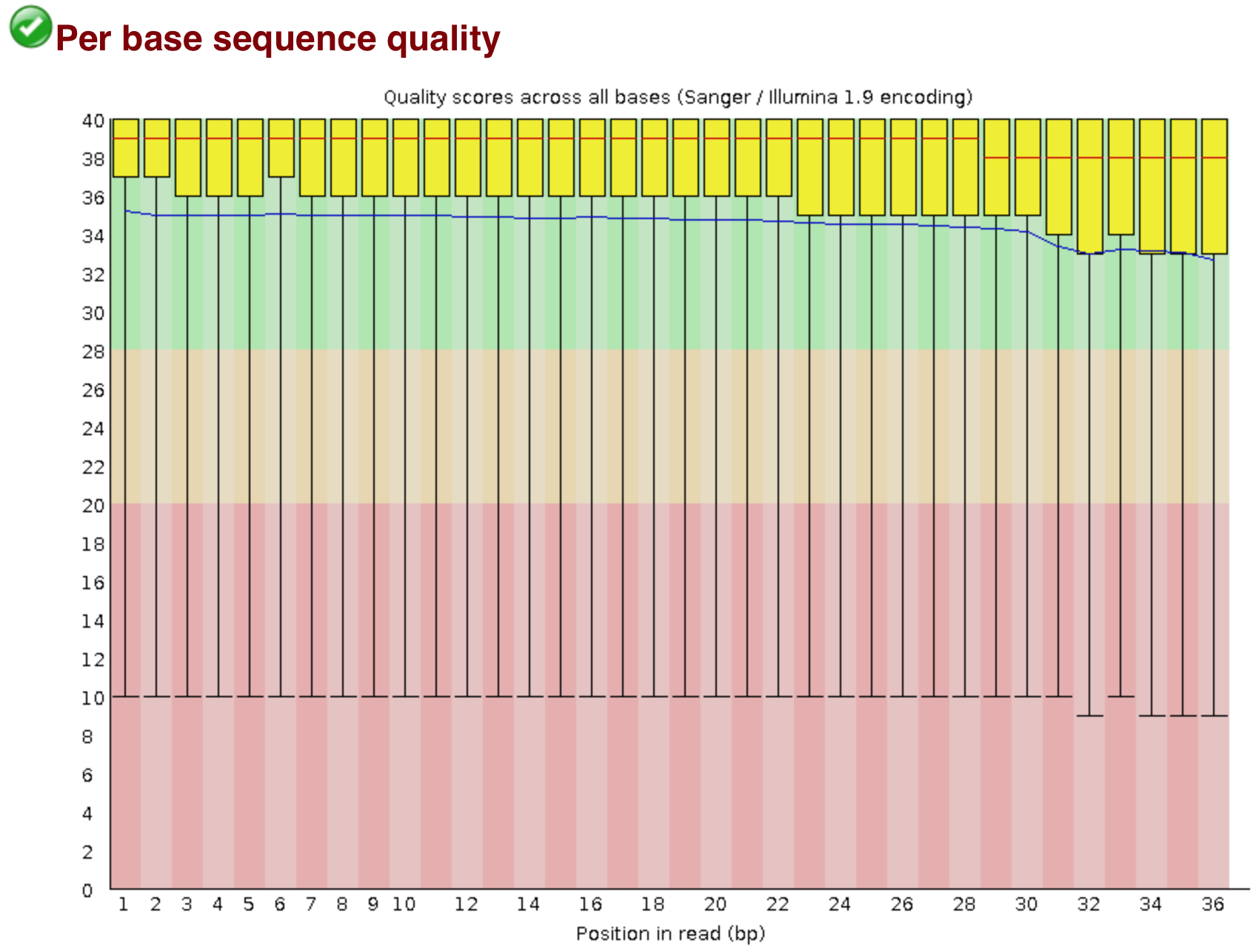
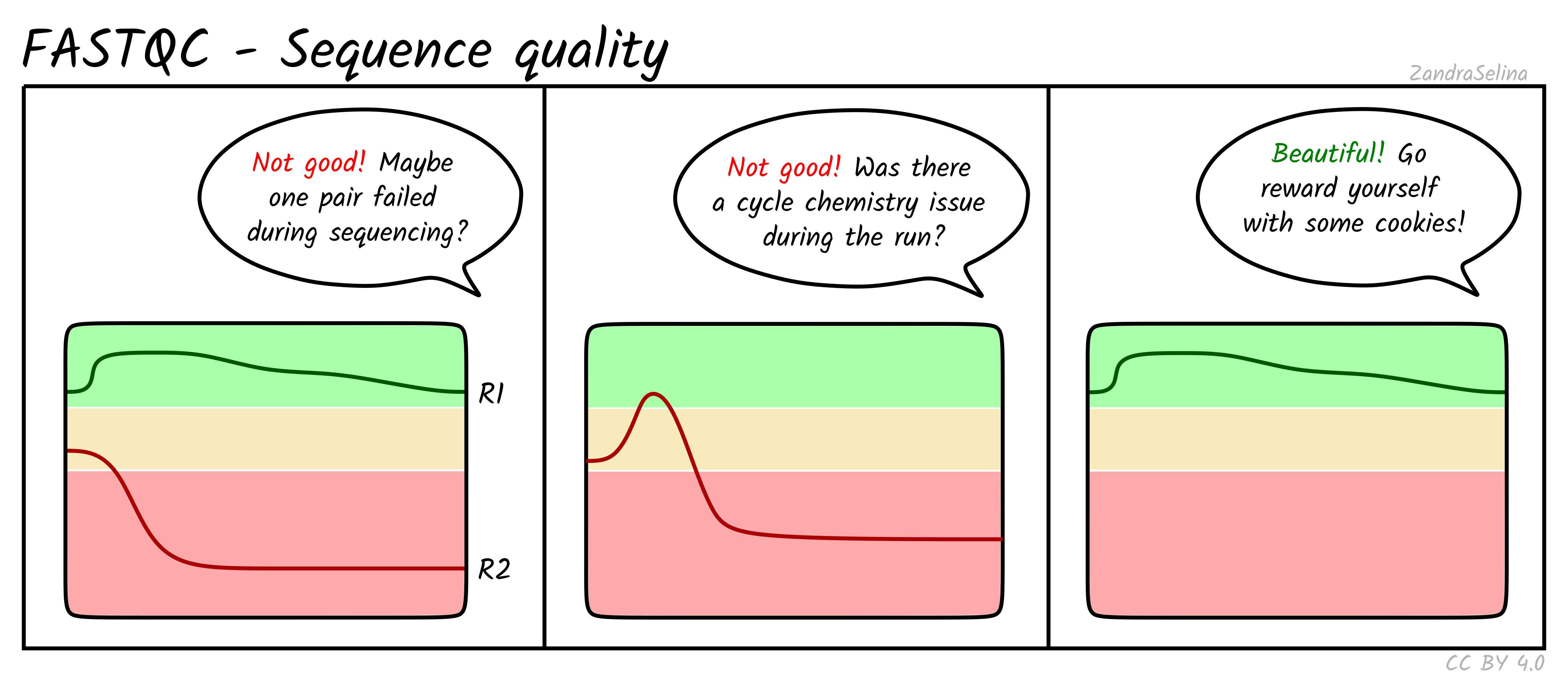
Quality control: Assessing FASTQC results | Introduction to RNA-Seq using high-performance computing - ARCHIVED

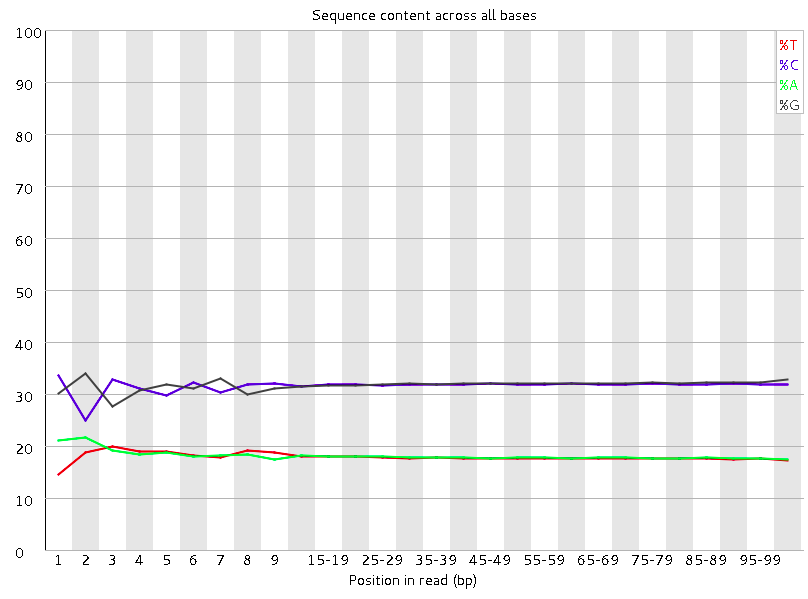
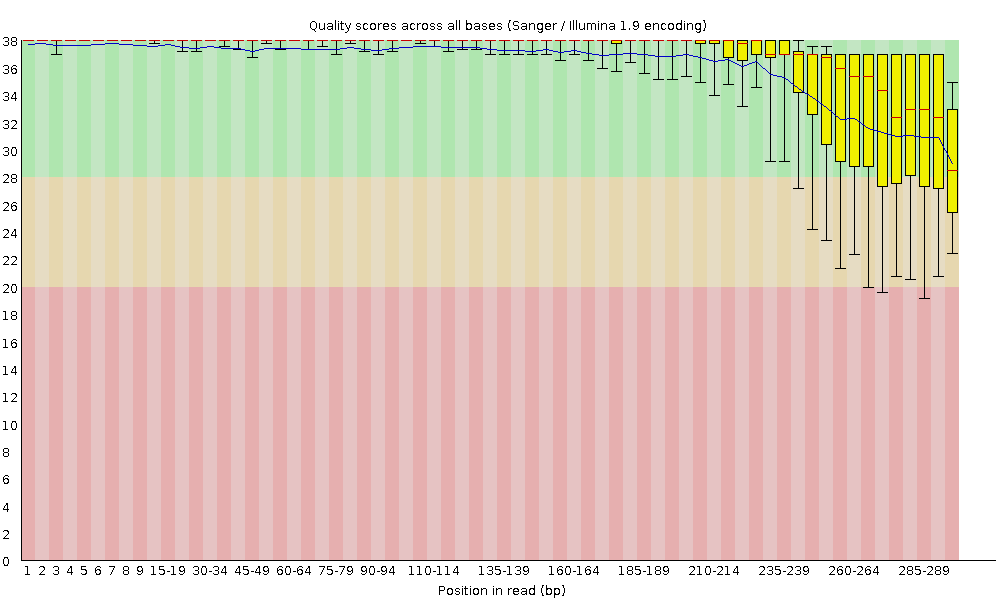
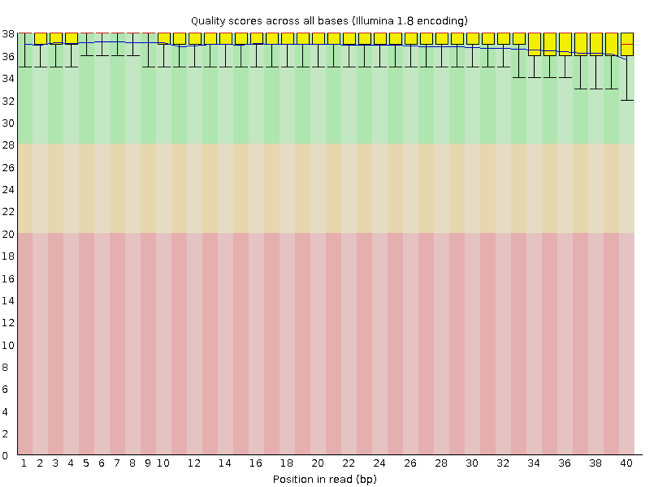
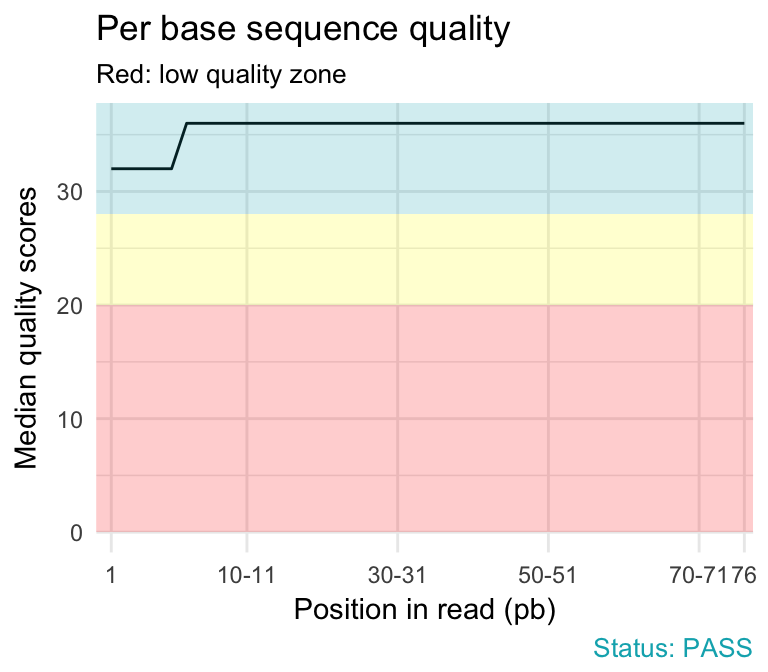
FastQC report of sequencing data. (A) Per base sequence quality; (B)... | Download Scientific Diagram

ClinQC quality control report generated by FASTQC. a Per base sequence... | Download Scientific Diagram