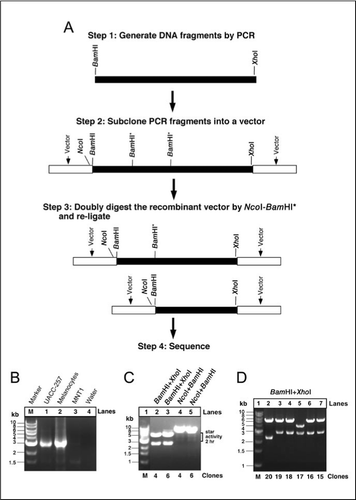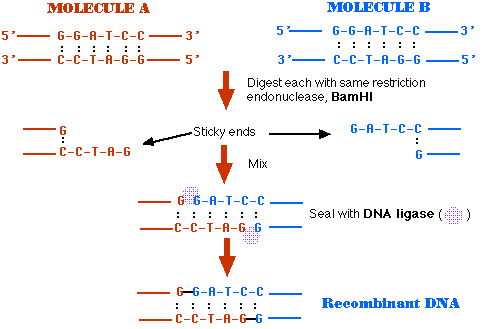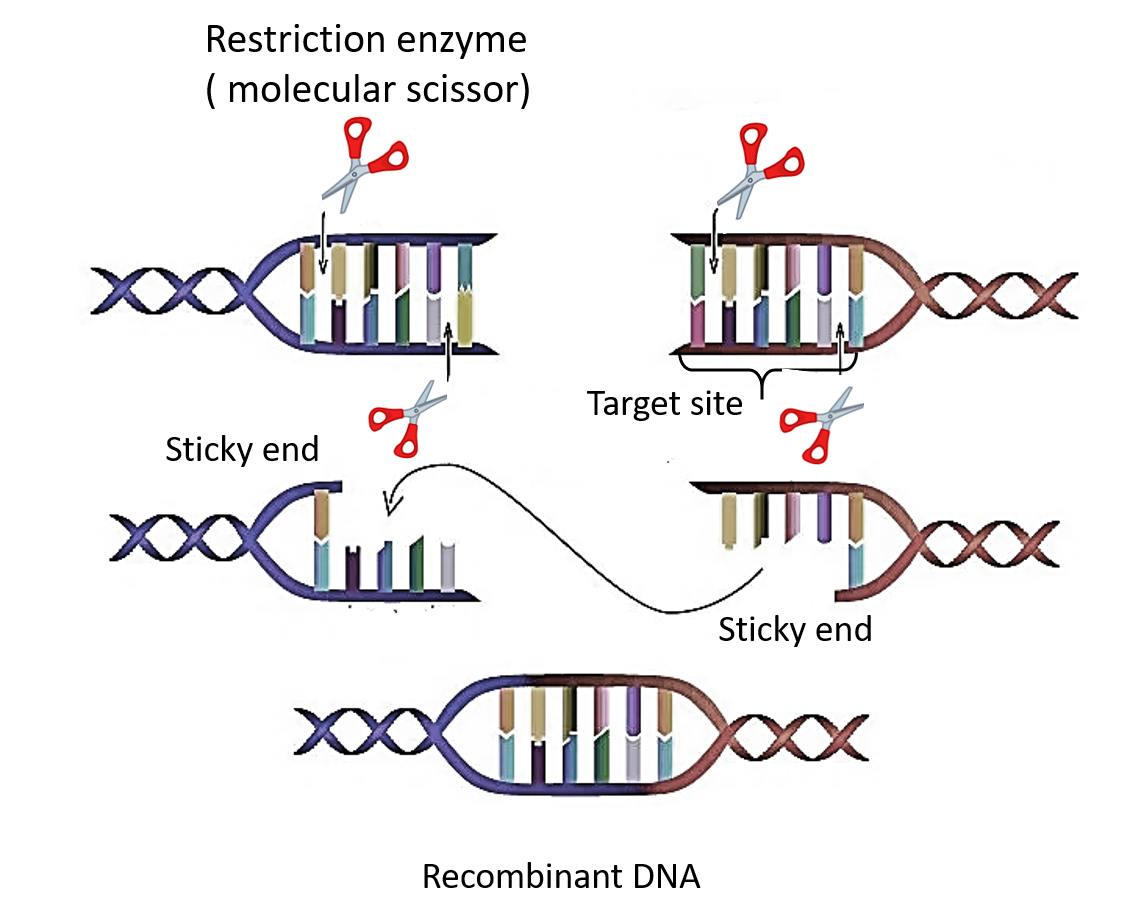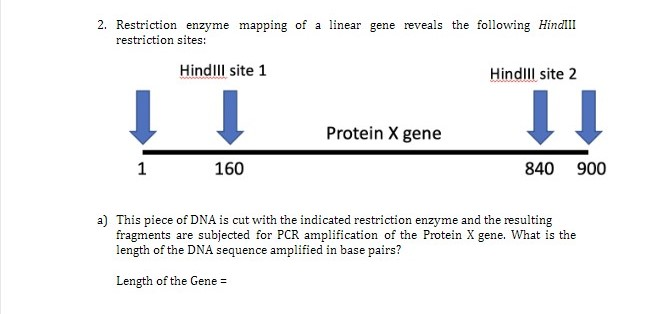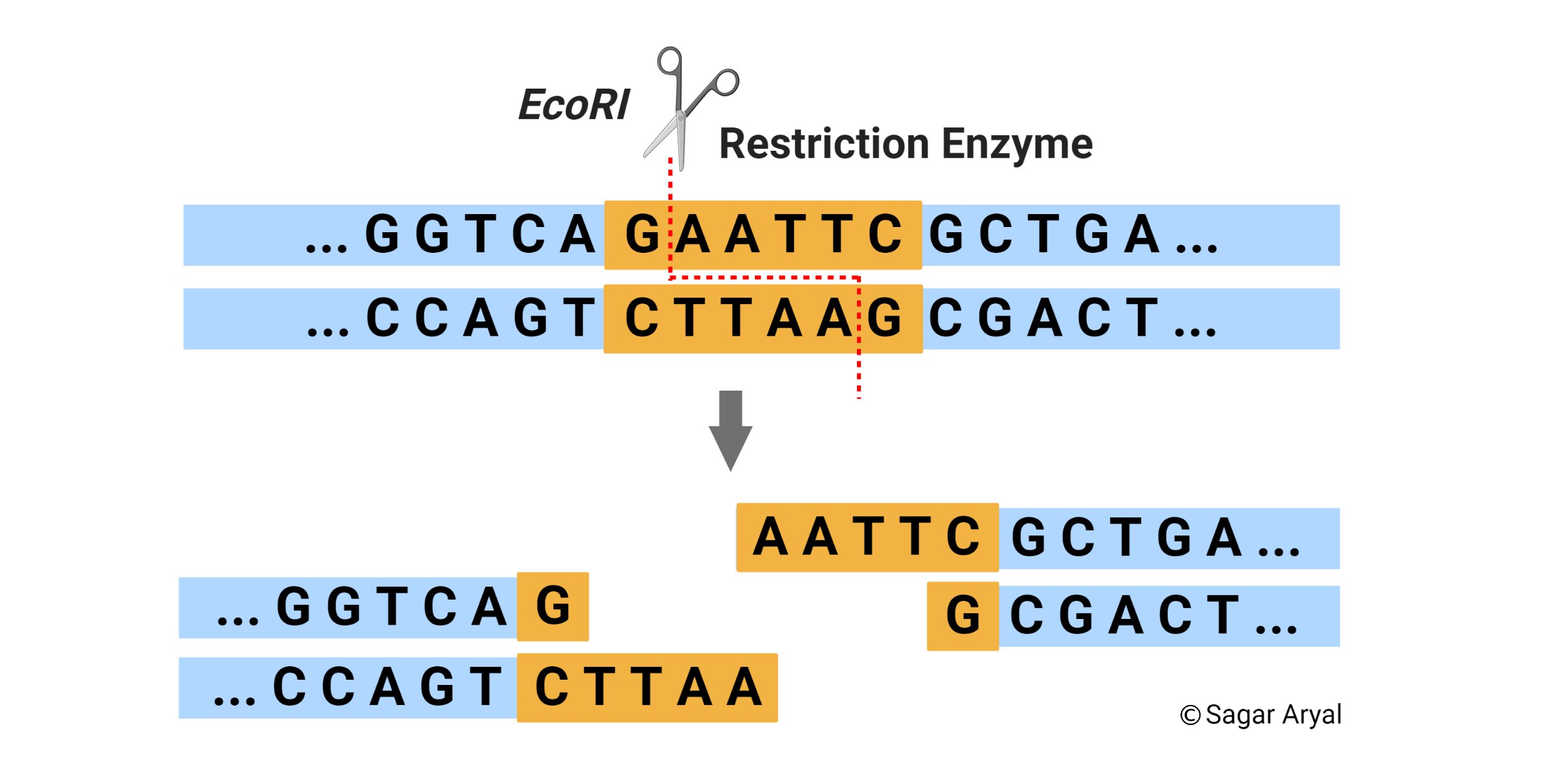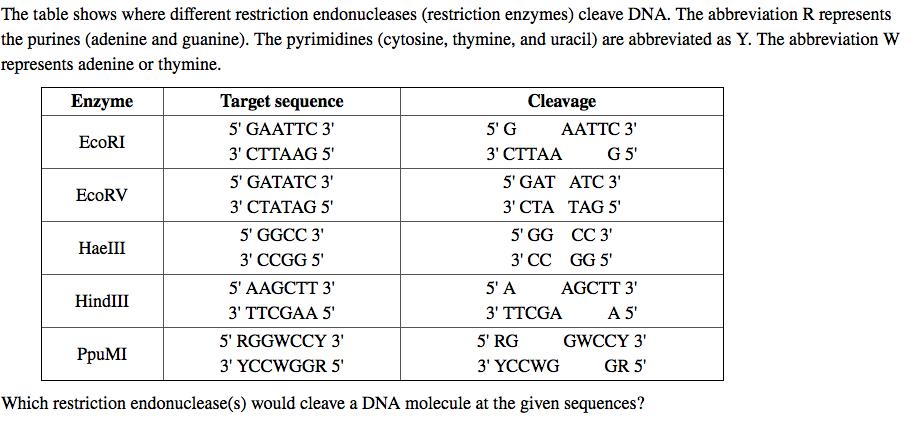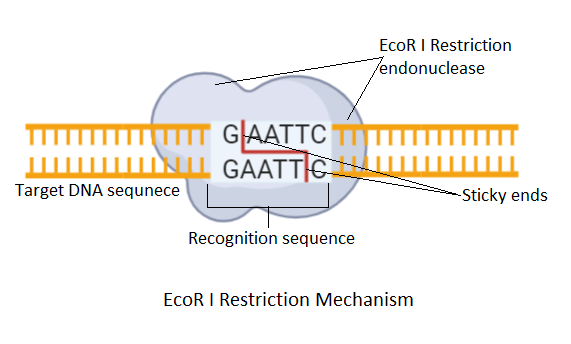
GAATTC is the restriction site, for which of the following restriction endonucleases?A. HindIIIB. EcoRIC. BamID. HaeIII

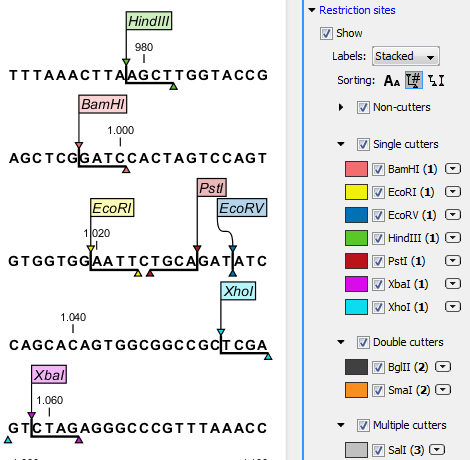
A linear piece of DNA is cleaved with the restriction enzymes HindIII and SmaI, each alone and in combination, to yield the fragments of the size shown below: HindIII: 1.0, 2.5, and

SOLVED: You have chosen to cut your DNA and plasmid with the HindIII restriction enzyme. This enzyme recognizes the sequence AAGCTT and makes a cut between the two A bases. This will