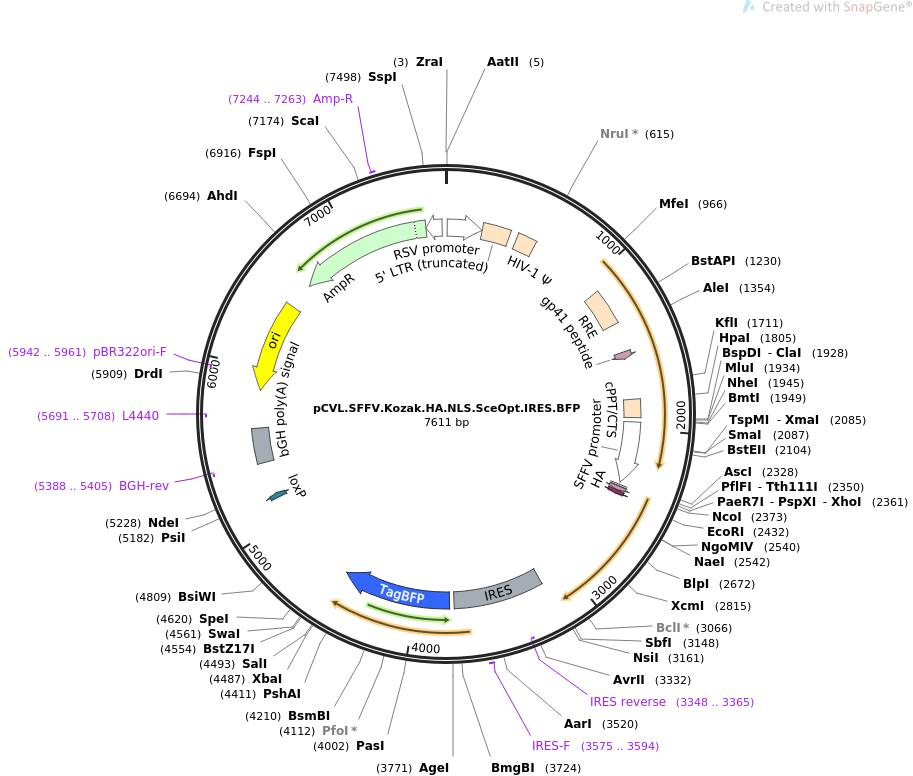
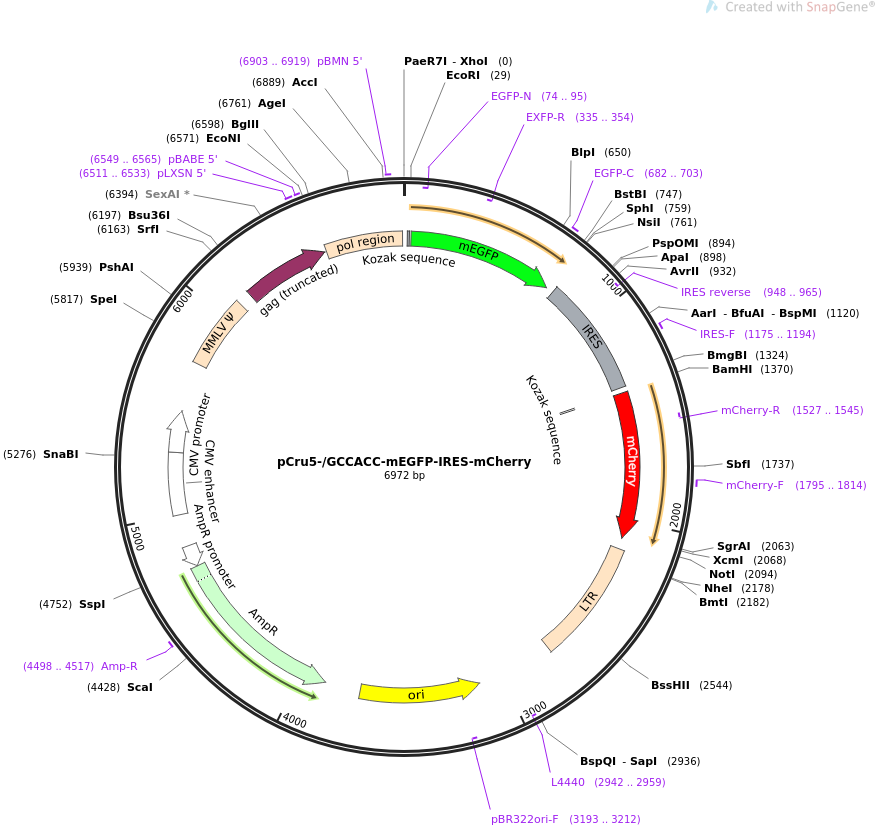
Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants | Microbial Cell Factories | Full Text

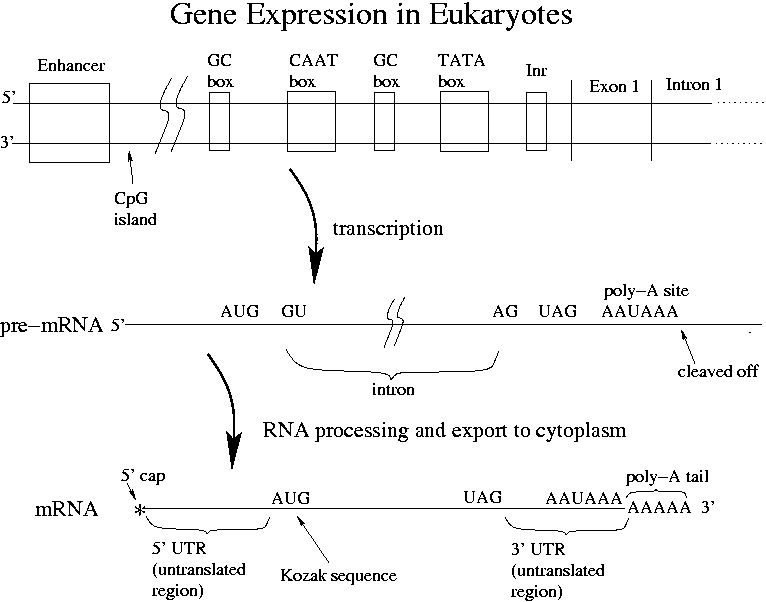
Bioinformatic analyses of mammalian 5'-UTR sequence properties of mRNAs predicts alternative translation initiation sites | BMC Bioinformatics | Full Text
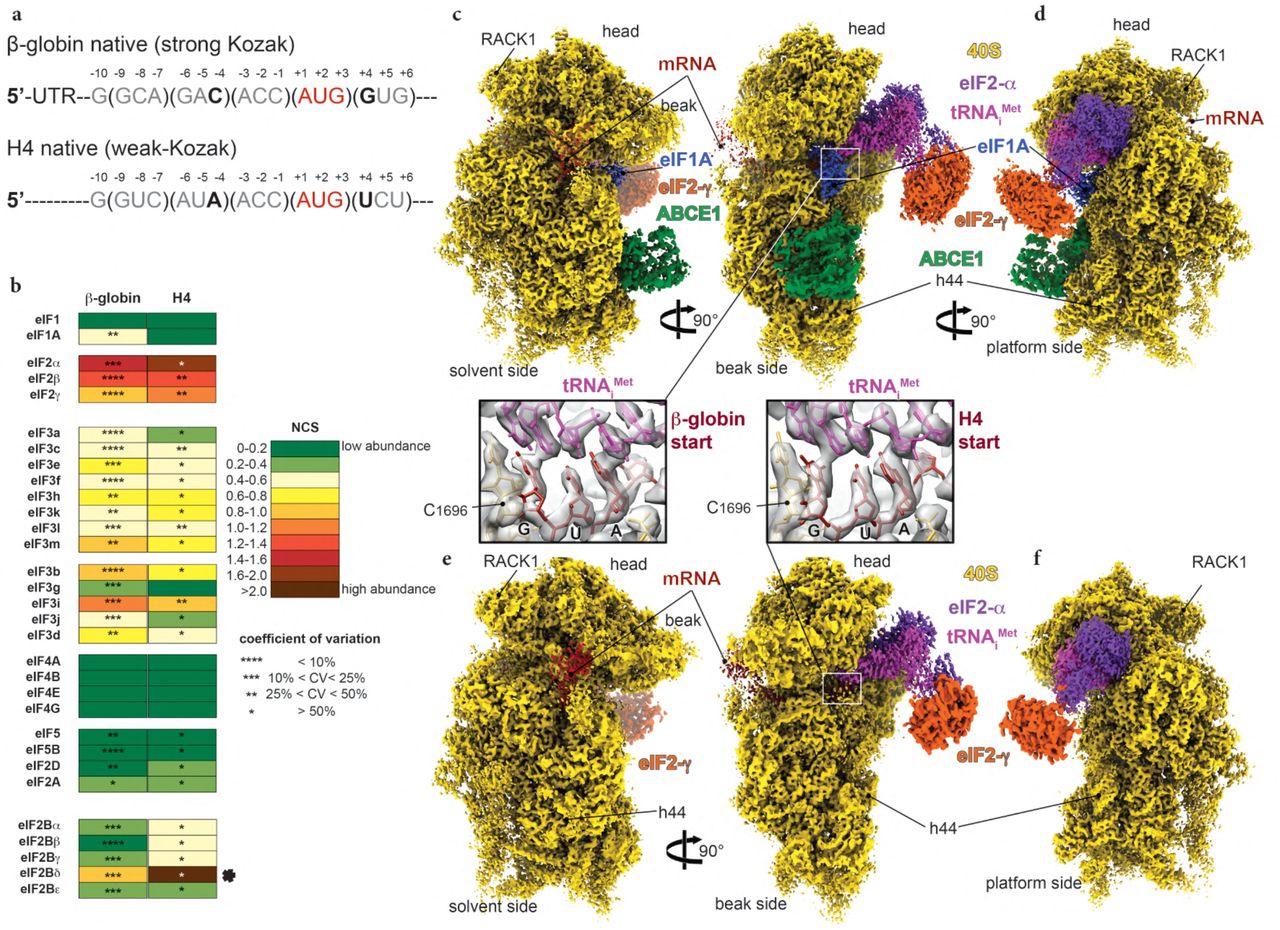
Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv
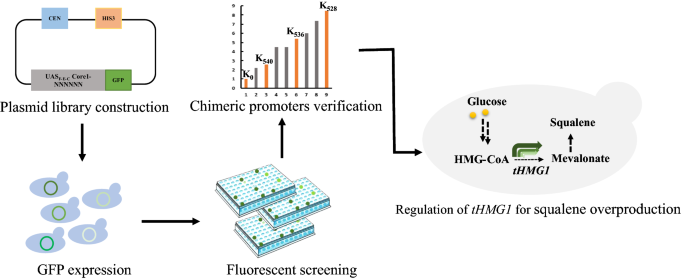
Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants | Microbial Cell Factories | Full Text

Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv

Quantitative analysis of mammalian translation initiation sites by FACS‐seq | Molecular Systems Biology
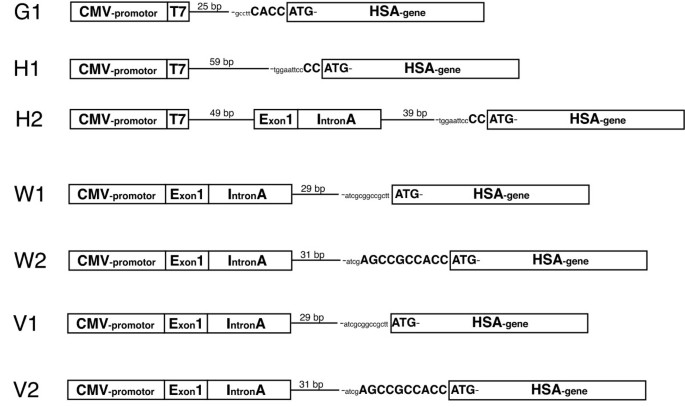
In vitro analysis of expression vectors for DNA vaccination of horses: the effect of a Kozak sequence | Acta Veterinaria Scandinavica | Full Text
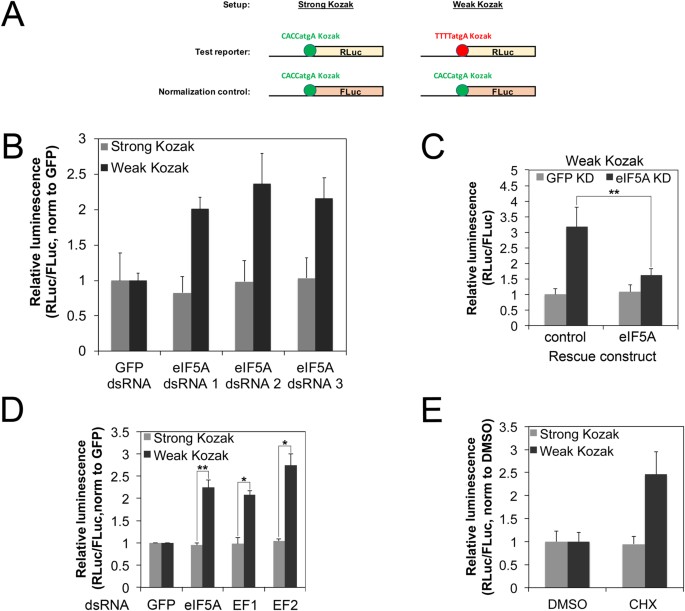
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports
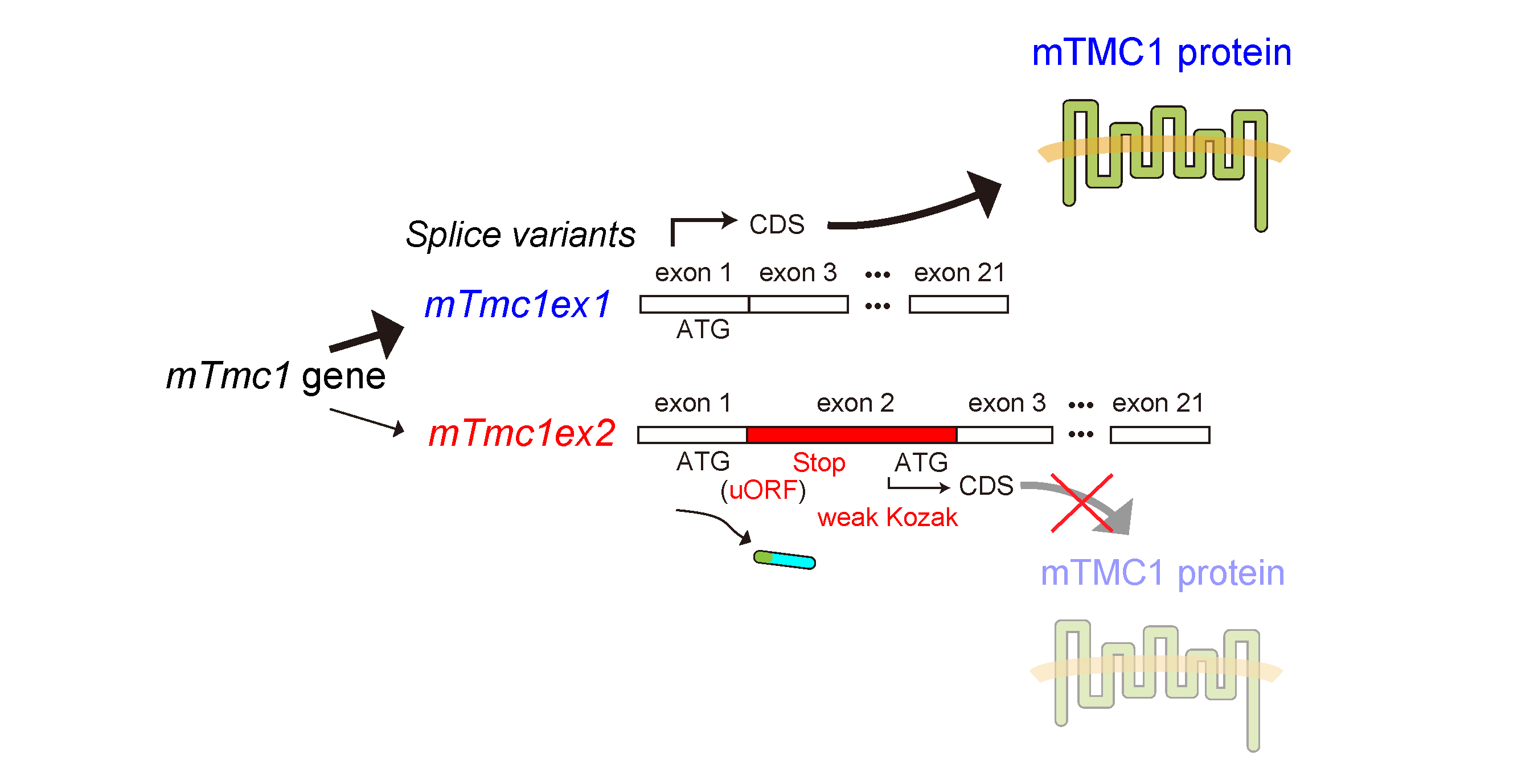
IJMS | Free Full-Text | A Mechanosensitive Channel, Mouse Transmembrane Channel-Like Protein 1 (mTMC1) Is Translated from a Splice Variant mTmc1ex1 but Not from the Other Variant mTmc1ex2
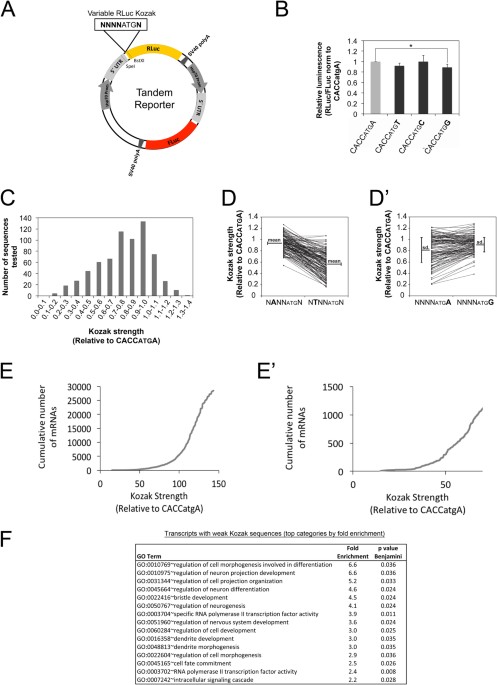
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports

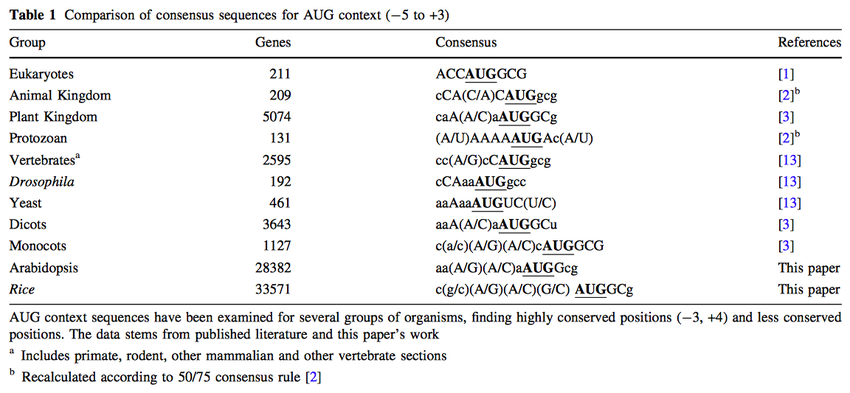
Relationship between consensus Kozak sequence at the true start ATG for... | Download Scientific Diagram

IRES-Dependent Second Gene Expression Is Significantly Lower Than Cap-Dependent First Gene Expression in a Bicistronic Vector: Molecular Therapy

Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv