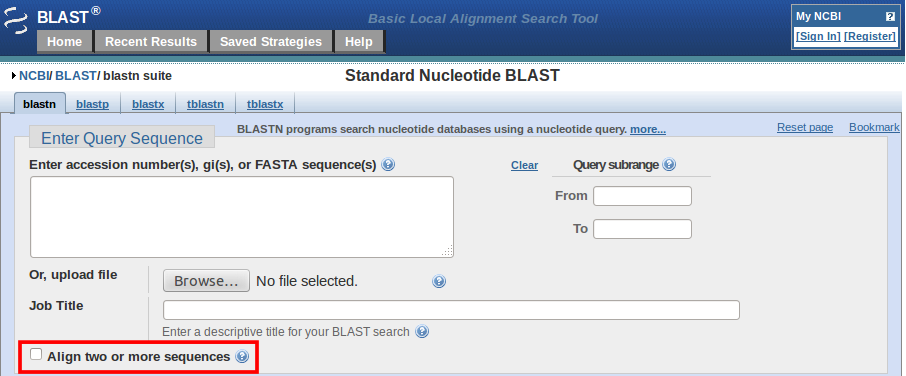
BLAST: Compare & identify sequences - NCBI Bioinformatics Resources: An Introduction - Library Guides at UC Berkeley

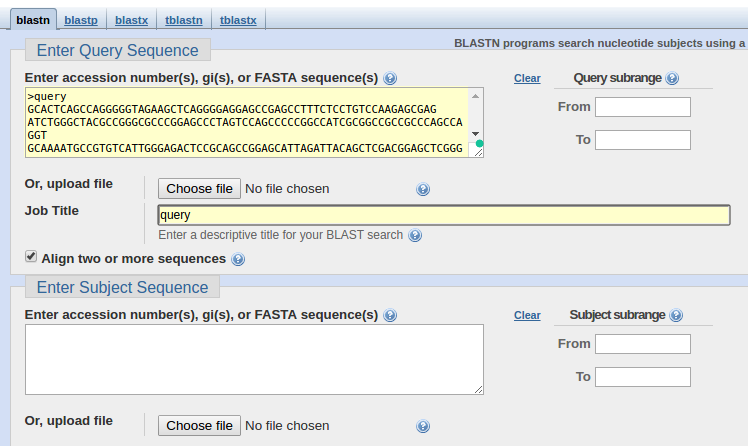
Figure 1, The new Blast 2 Sequences option on the Basic nucleotide BLAST form - NCBI News - NCBI Bookshelf

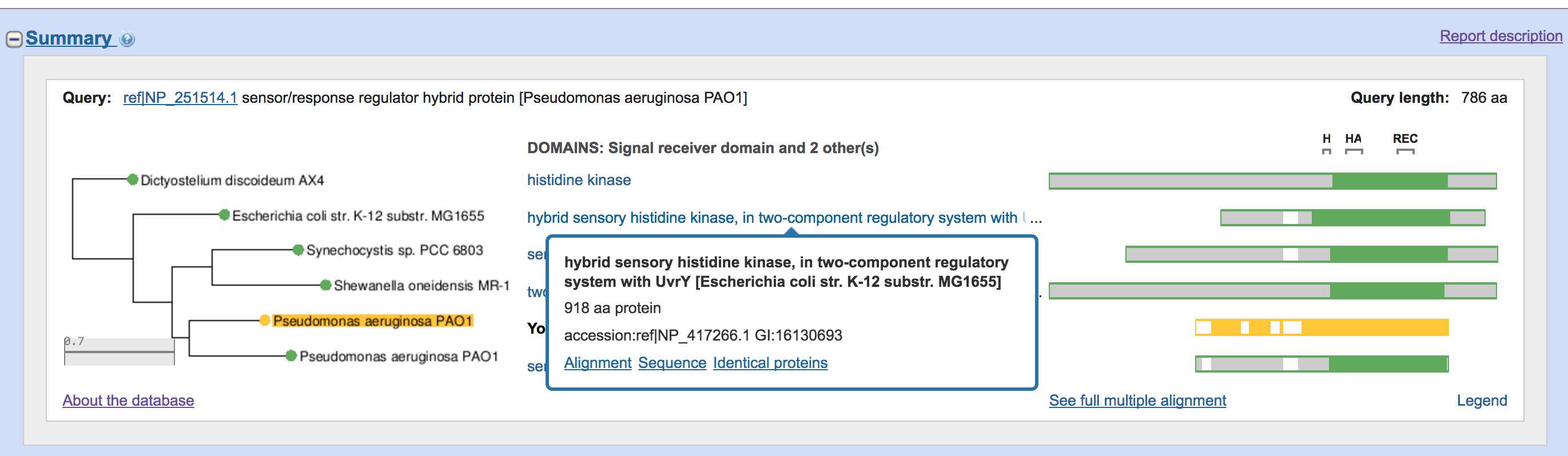
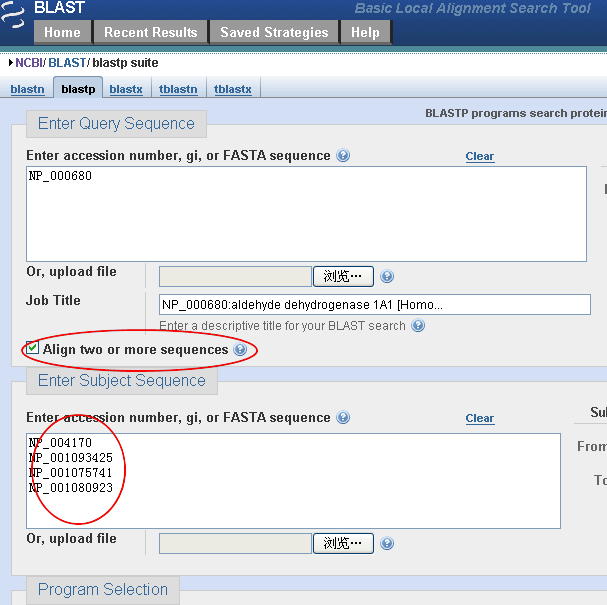
Using the NCBI BLAST Interface For Identifying Homologous Sequences In Differentiating Proteins Inside Arabidopsis Thaliana | by Karthik Mittal | Analytics Vidhya | Medium

6 NCBI BLAST pairwise alignment. The two partial sequences depicted in... | Download Scientific Diagram

Figure 2, The expanded Dot Matrix view from Blast 2 Sequences showing the alignment of two Salmonella enterica subsp. enterica genome sequences (serovar Heidelberg str. SL476, accession NC_001083 and serovar Typhi Ty2,
![PDF] BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. | Semantic Scholar PDF] BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/92dffee93794394dd7d1b2db3dcb69177e0560cb/3-Figure1-1.png)