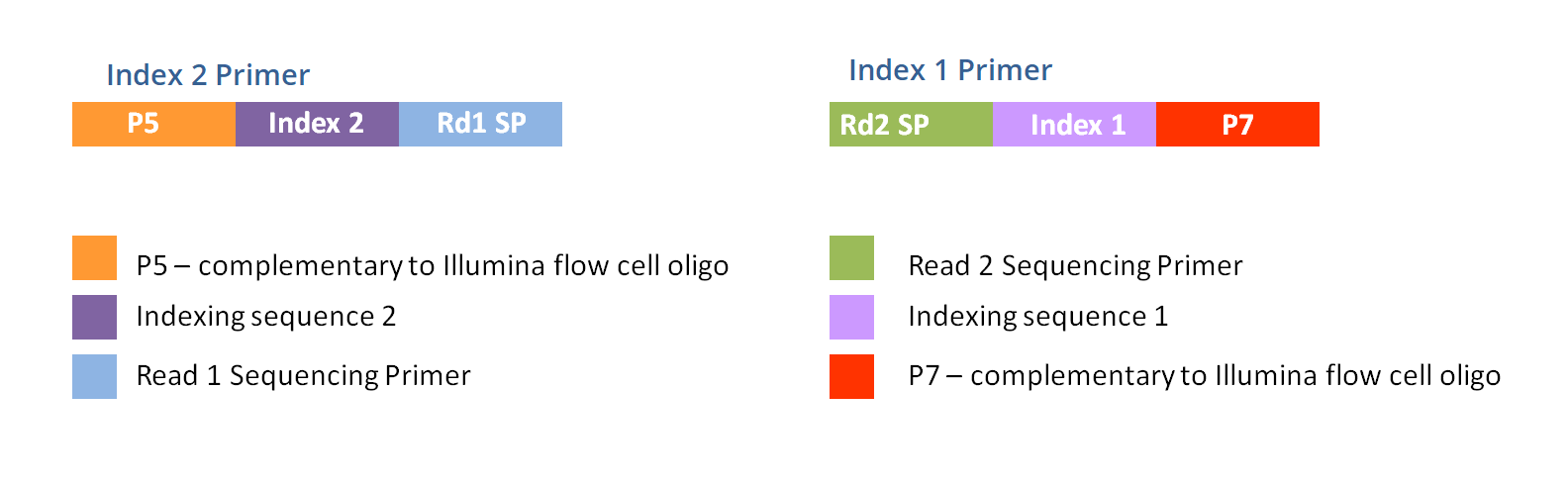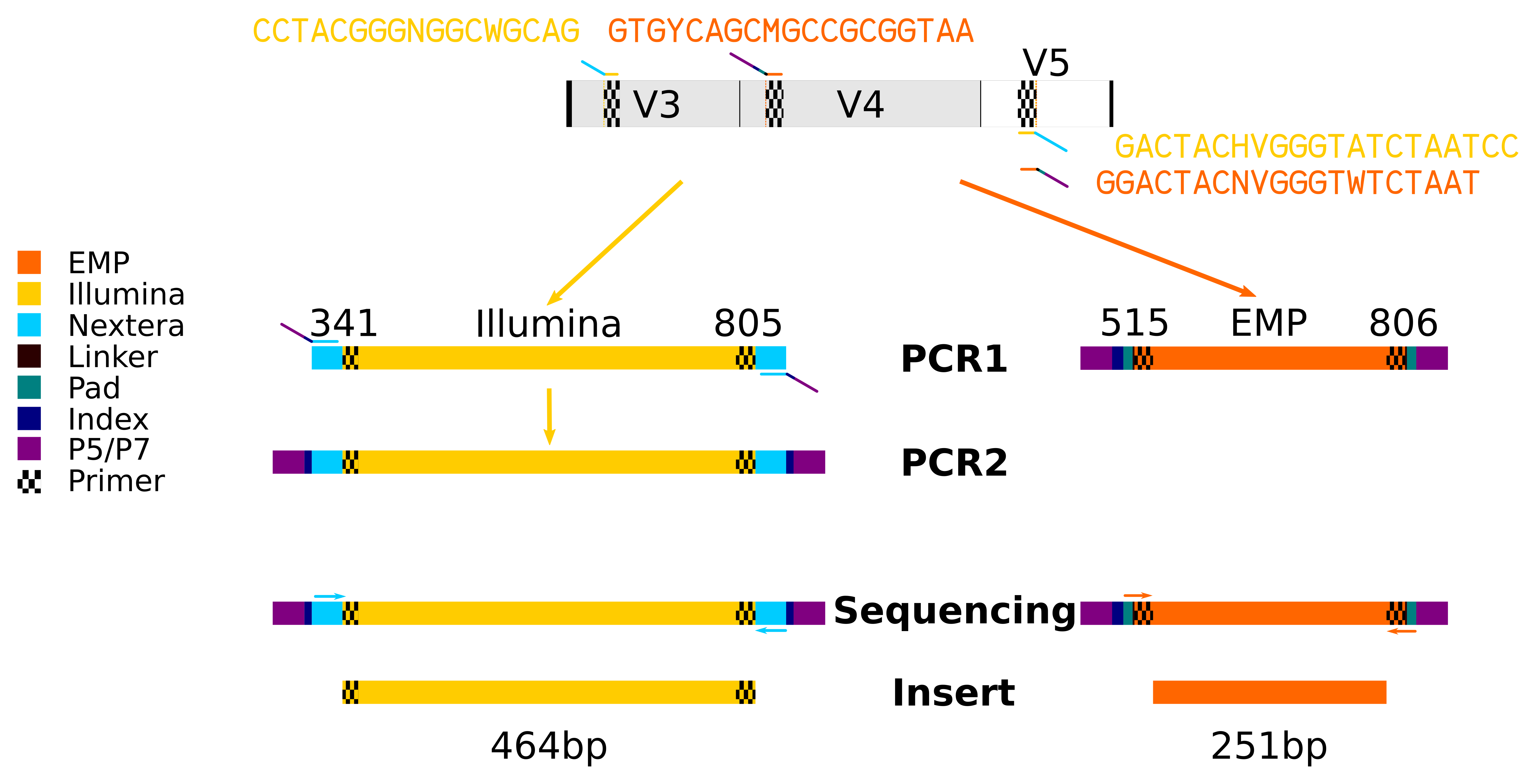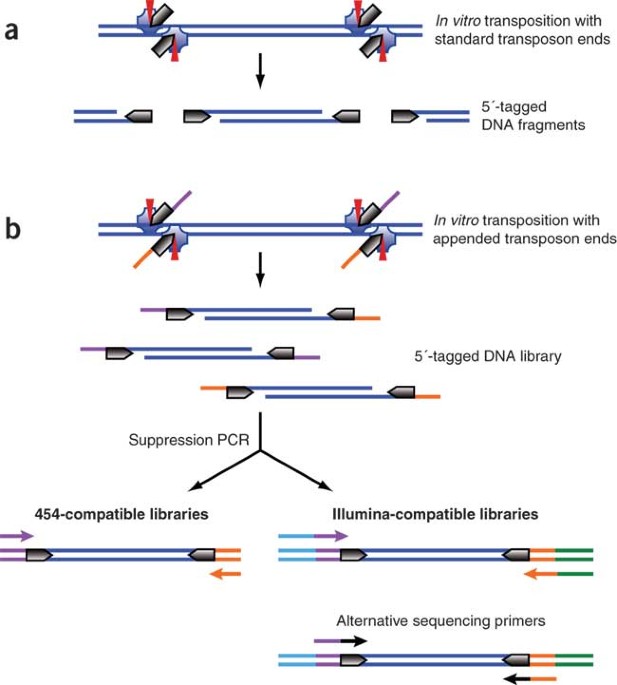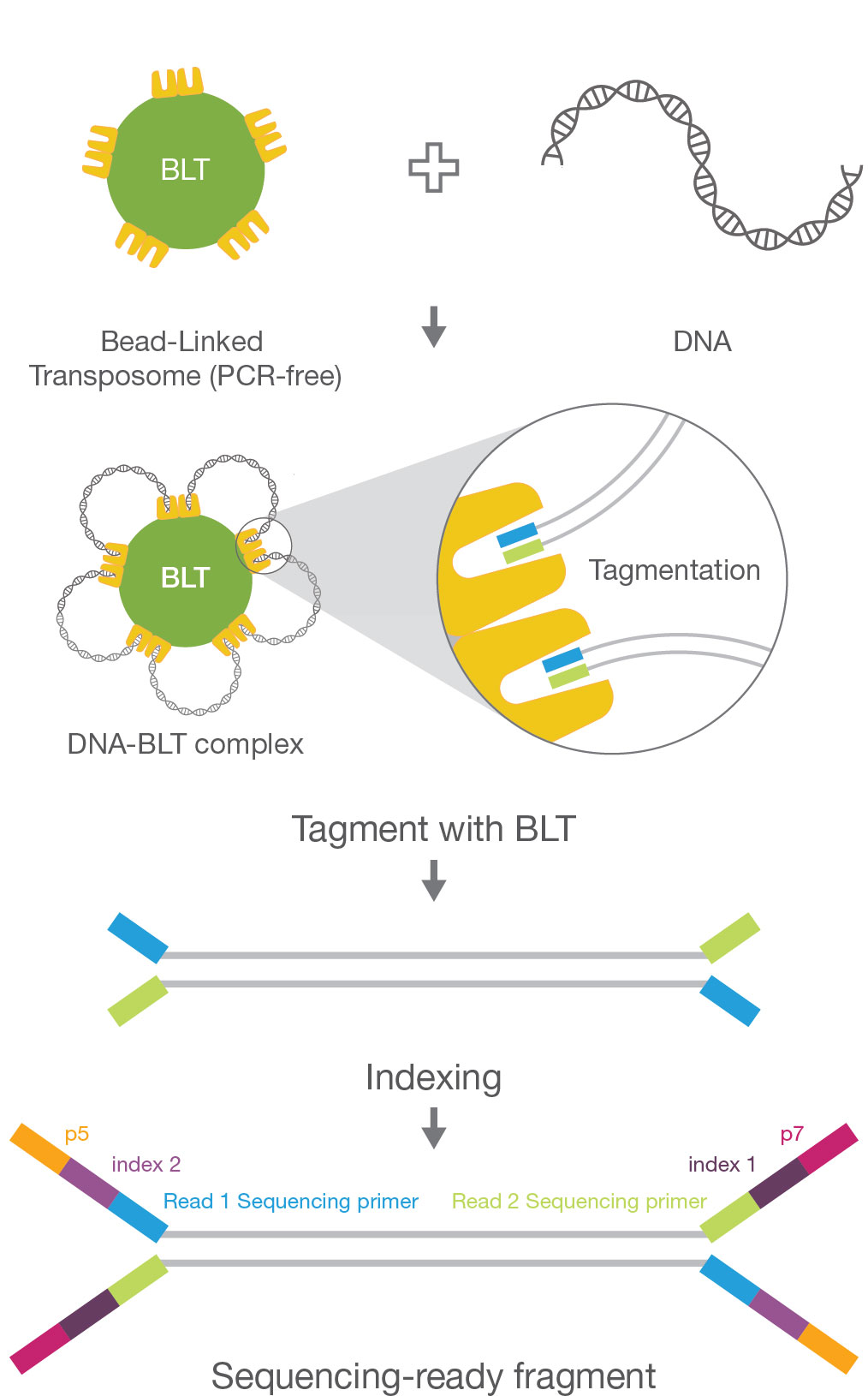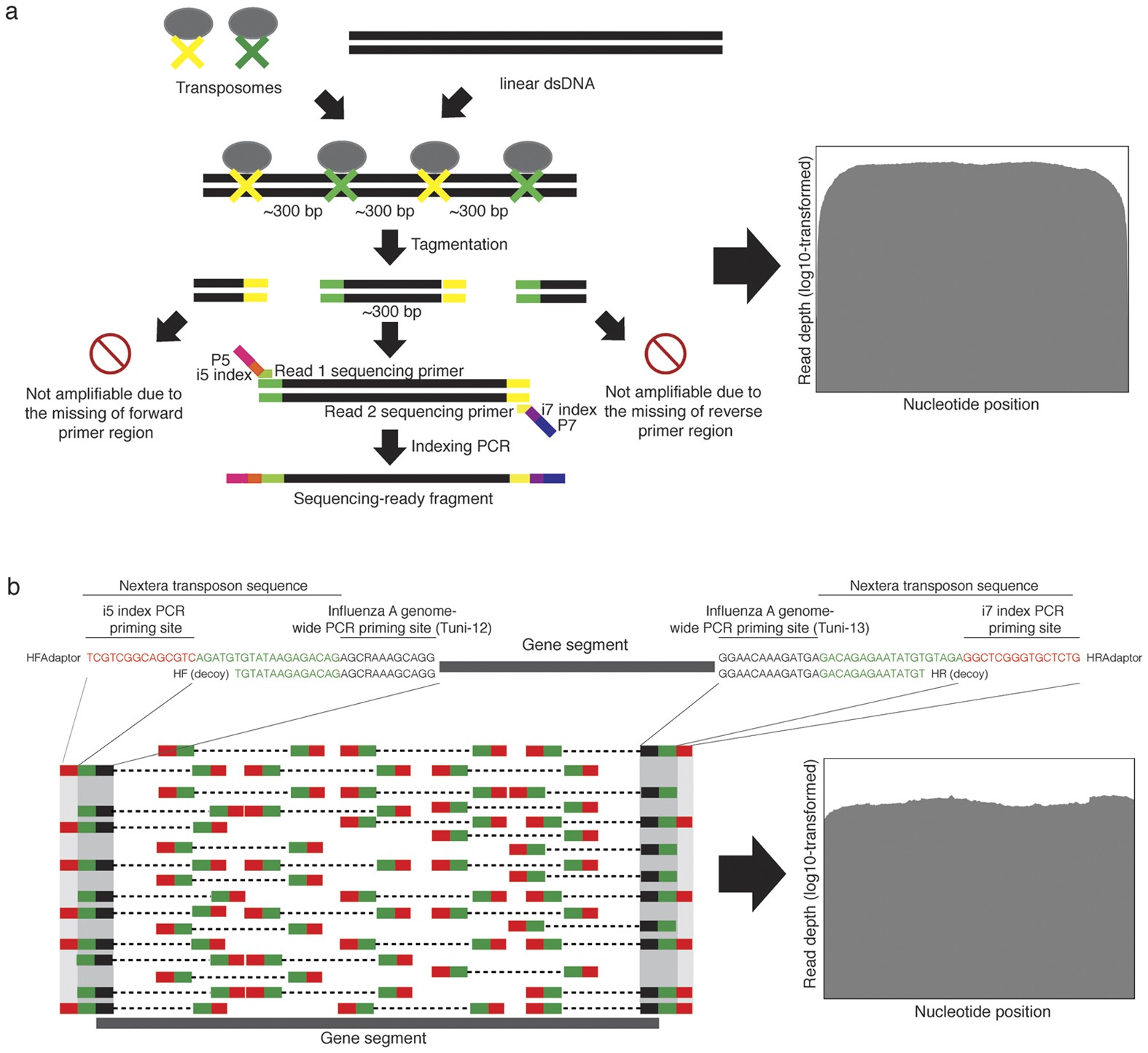
Contamination-controlled high-throughput whole genome sequencing for influenza A viruses using the MiSeq sequencer | Scientific Reports
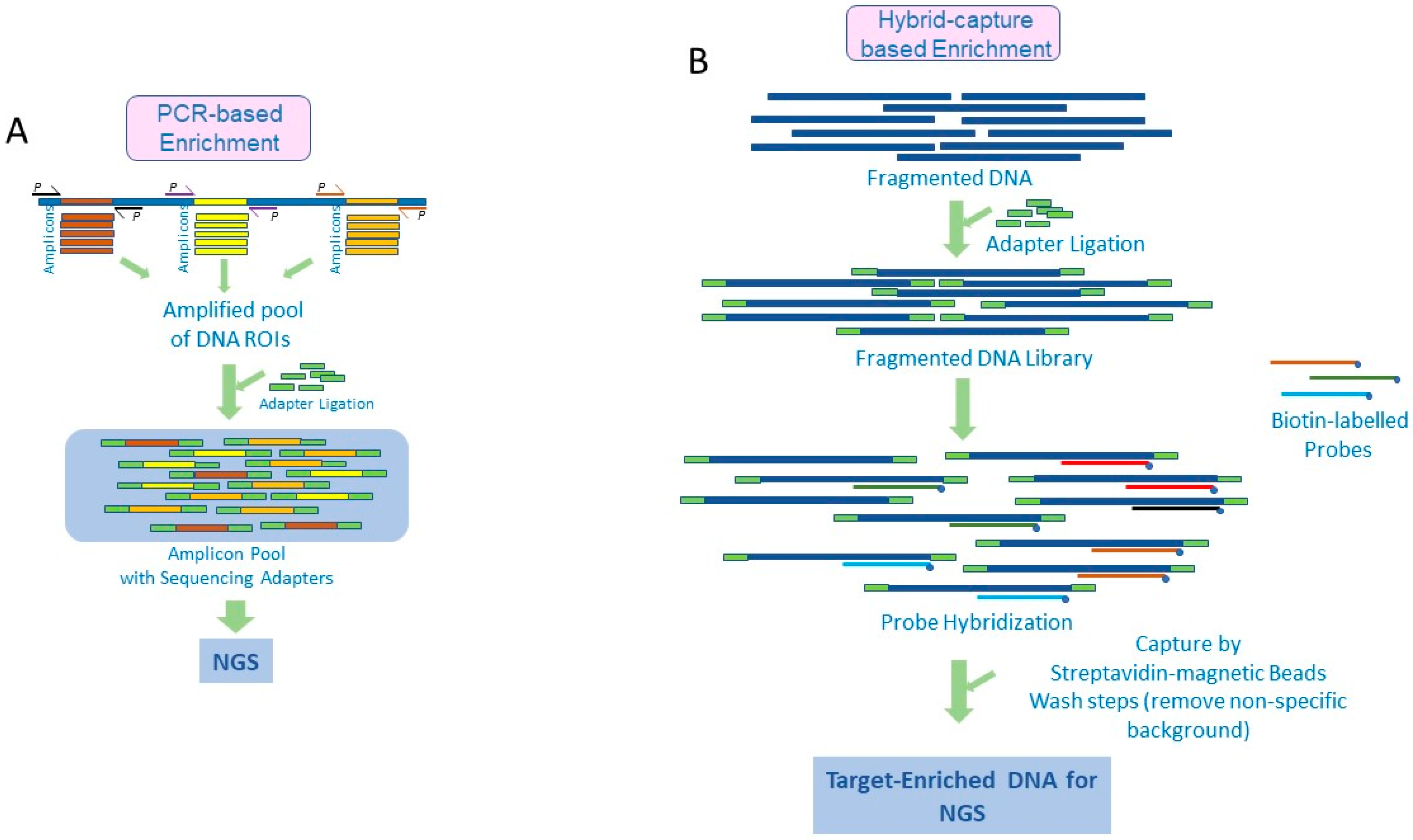
Diagnostics | Free Full-Text | Target Enrichment Approaches for Next-Generation Sequencing Applications in Oncology

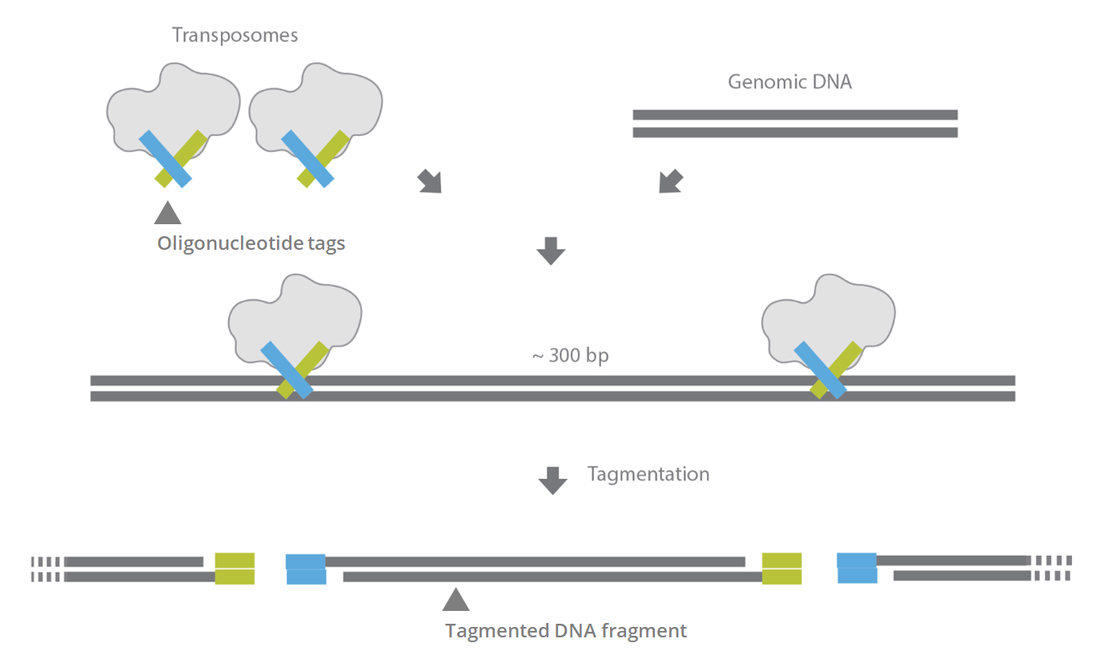
Library preparation for next generation sequencing: A review of automation strategies - ScienceDirect
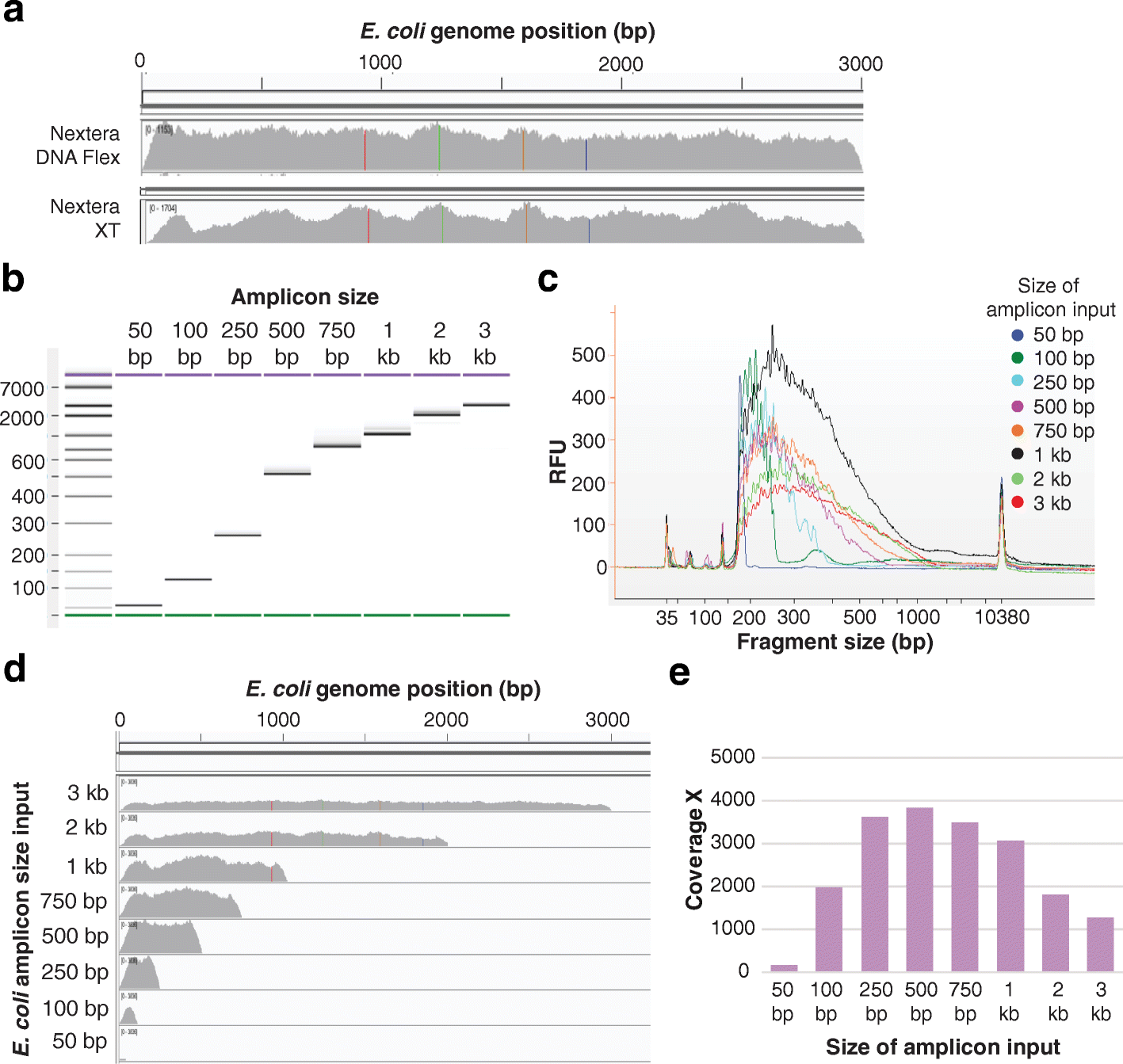


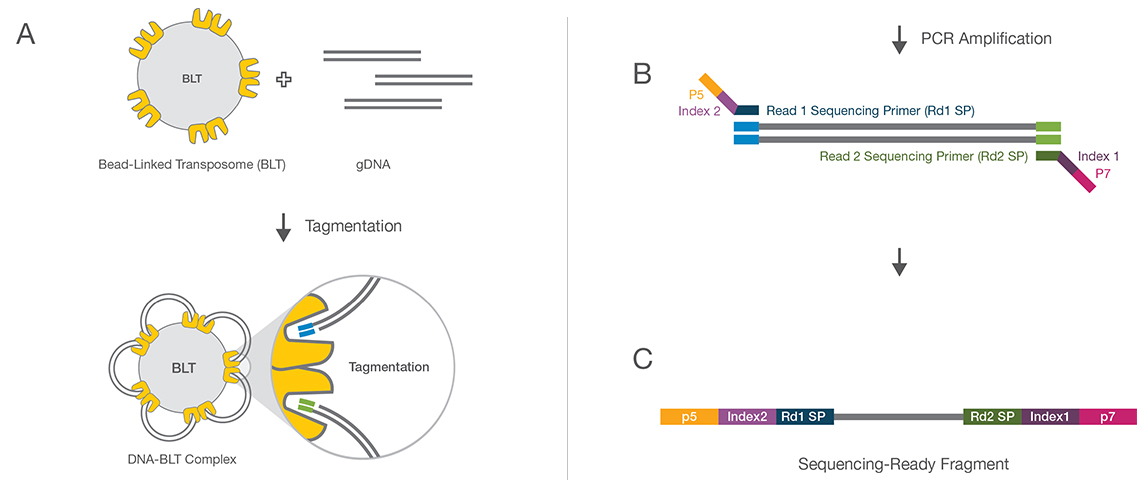
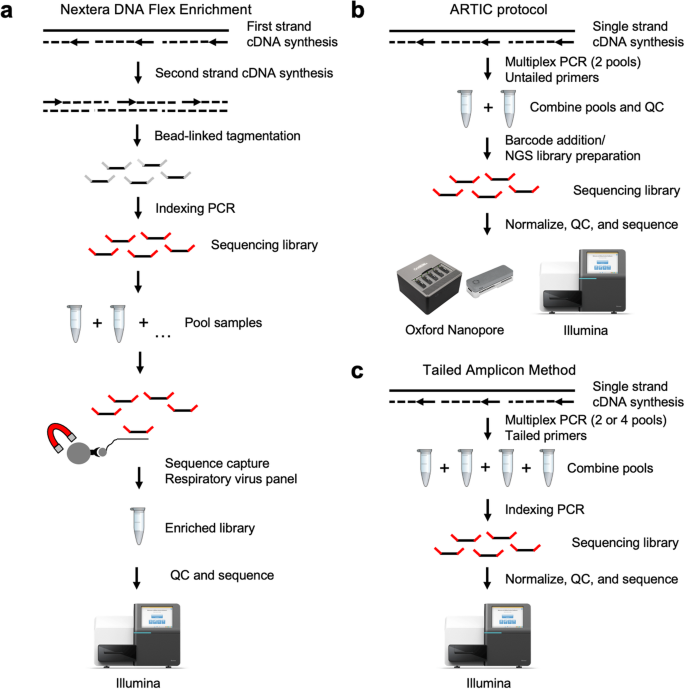

![PDF] Data Processing of Nextera Mate Pair Reads on Illumina Sequencing Platforms | Semantic Scholar PDF] Data Processing of Nextera Mate Pair Reads on Illumina Sequencing Platforms | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/6979583dd405728ade1fce397fa6d102a941394c/1-Figure1-1.png)