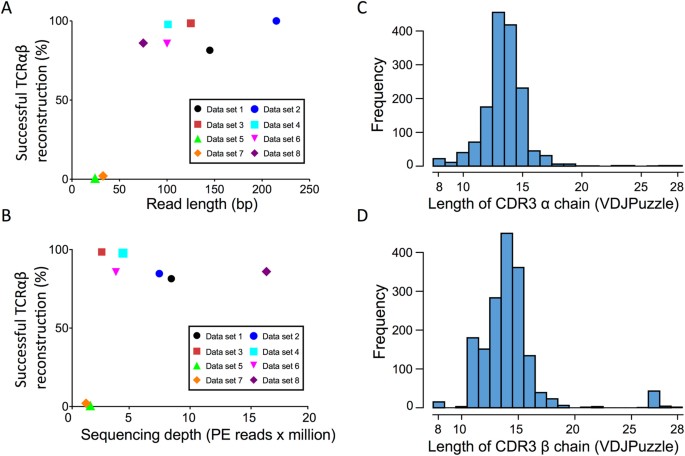
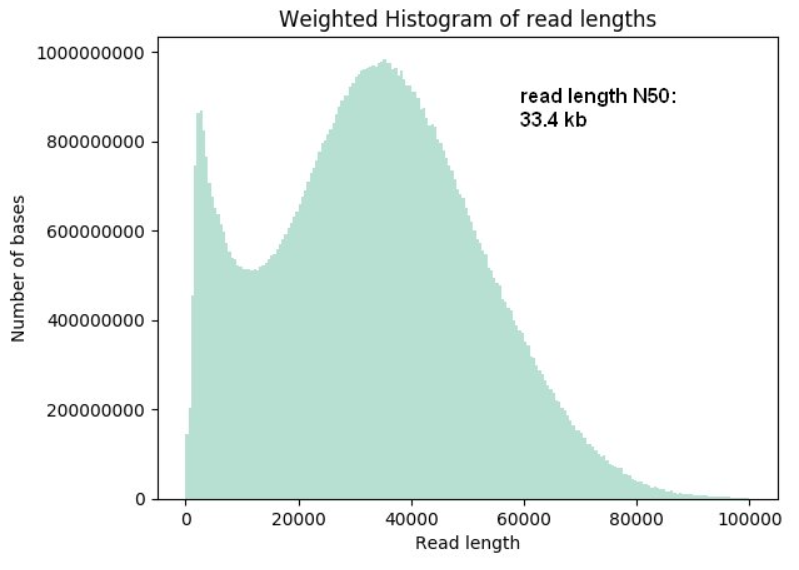
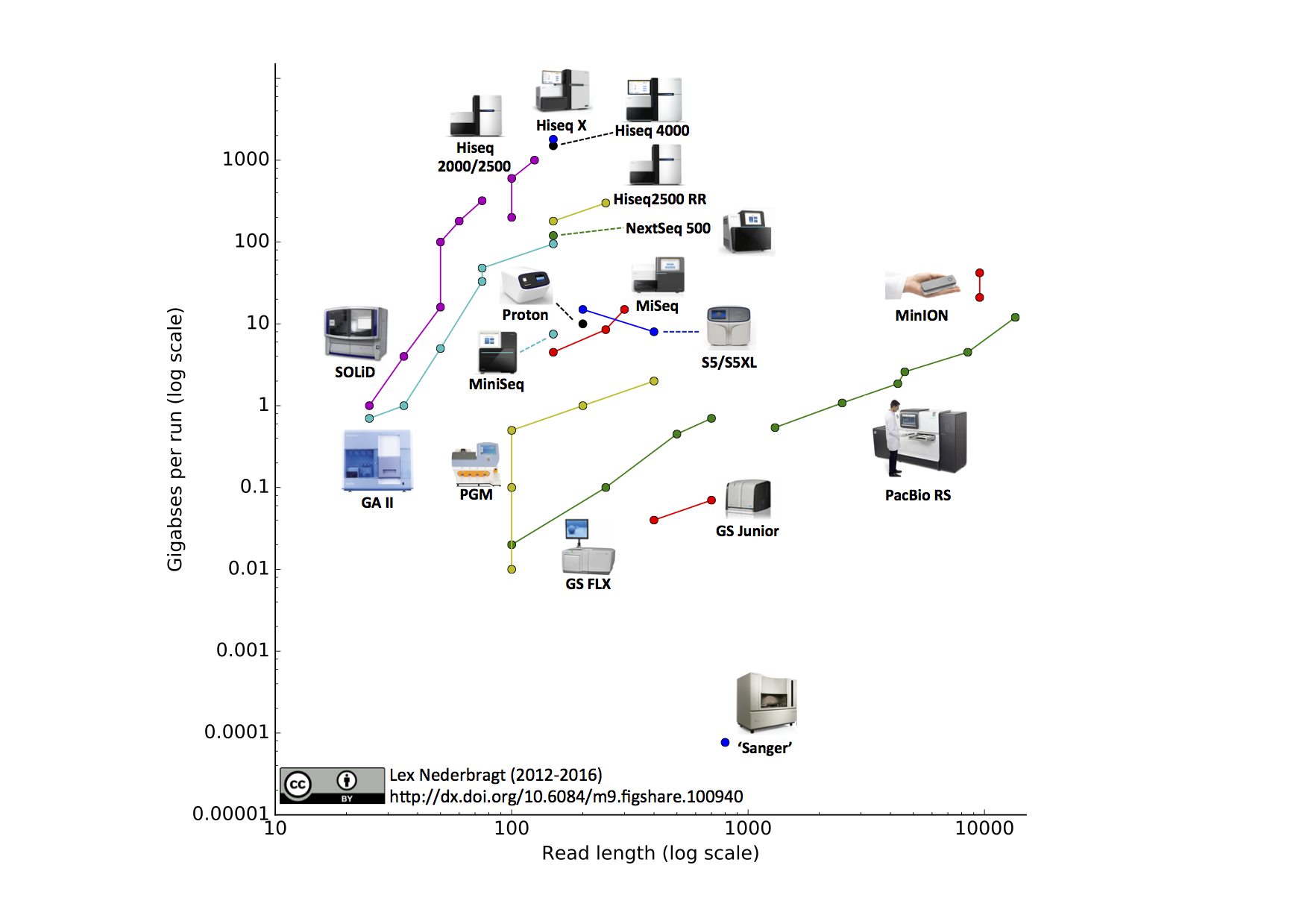
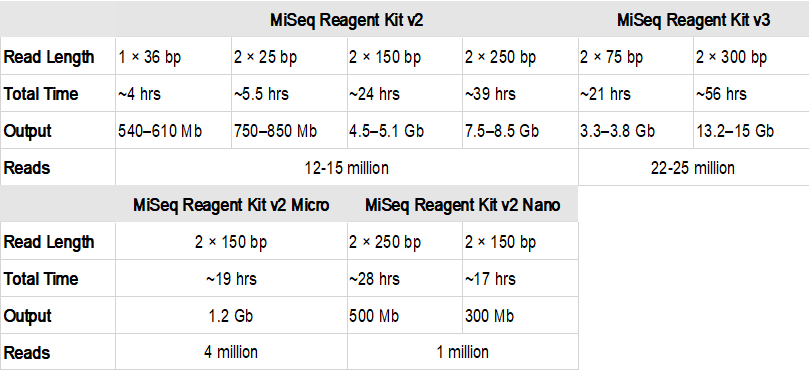
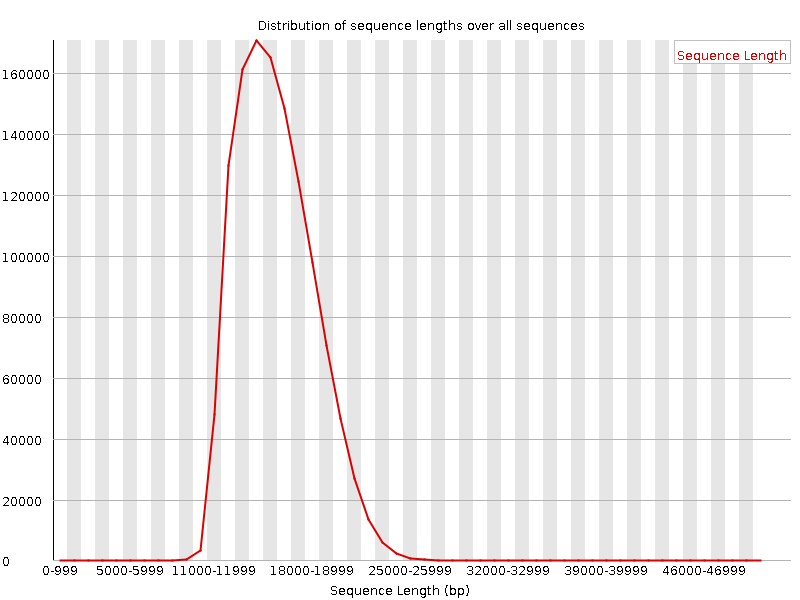
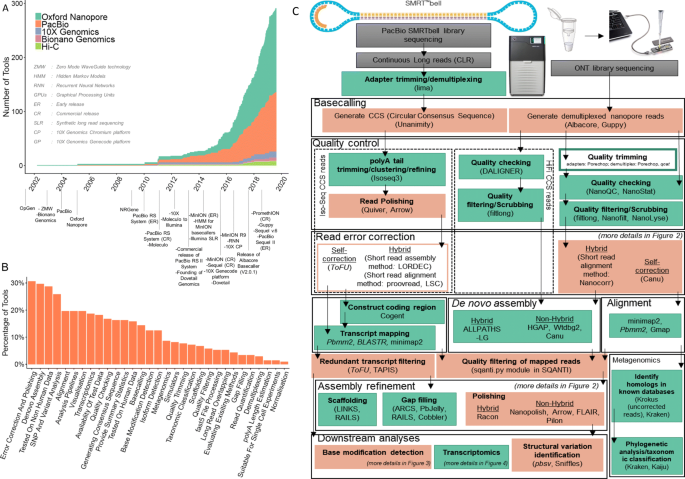
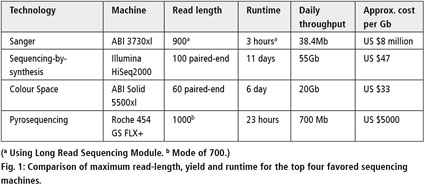
Longer and longer: DNA sequence of more than two million bases now achieved with nanopore sequencing.

From kilobases to "whales": a short history of ultra-long reads and high-throughput genome sequencing
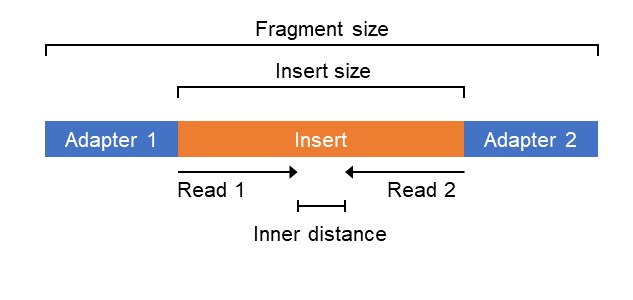
Compbio 020: Reads, fragments and inserts - what you need to know for understanding your sequencing data — Bad Grammar, Good Syntax

Oxford Nanopore Technology: A Promising Long-Read Sequencing Platform To Study Exon Connectivity and Characterize Isoforms of Complex Genes | Semantic Scholar