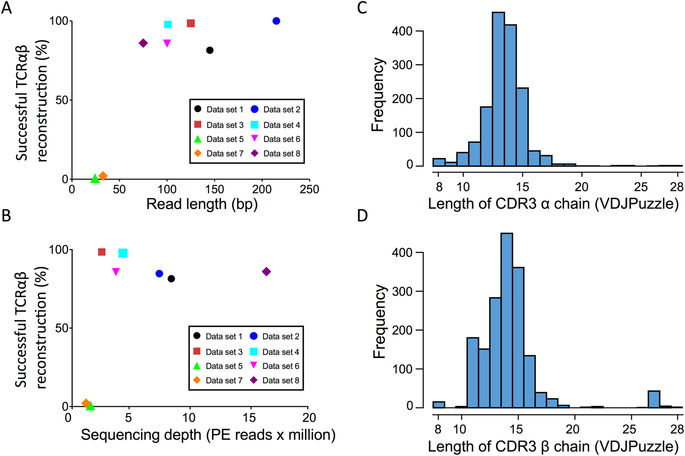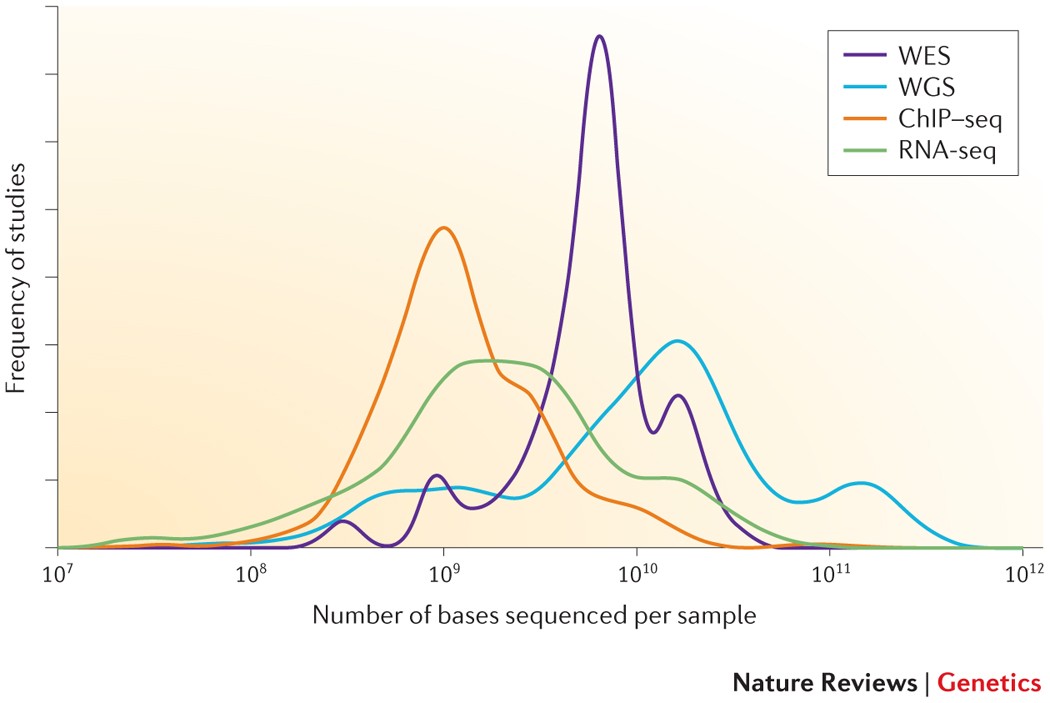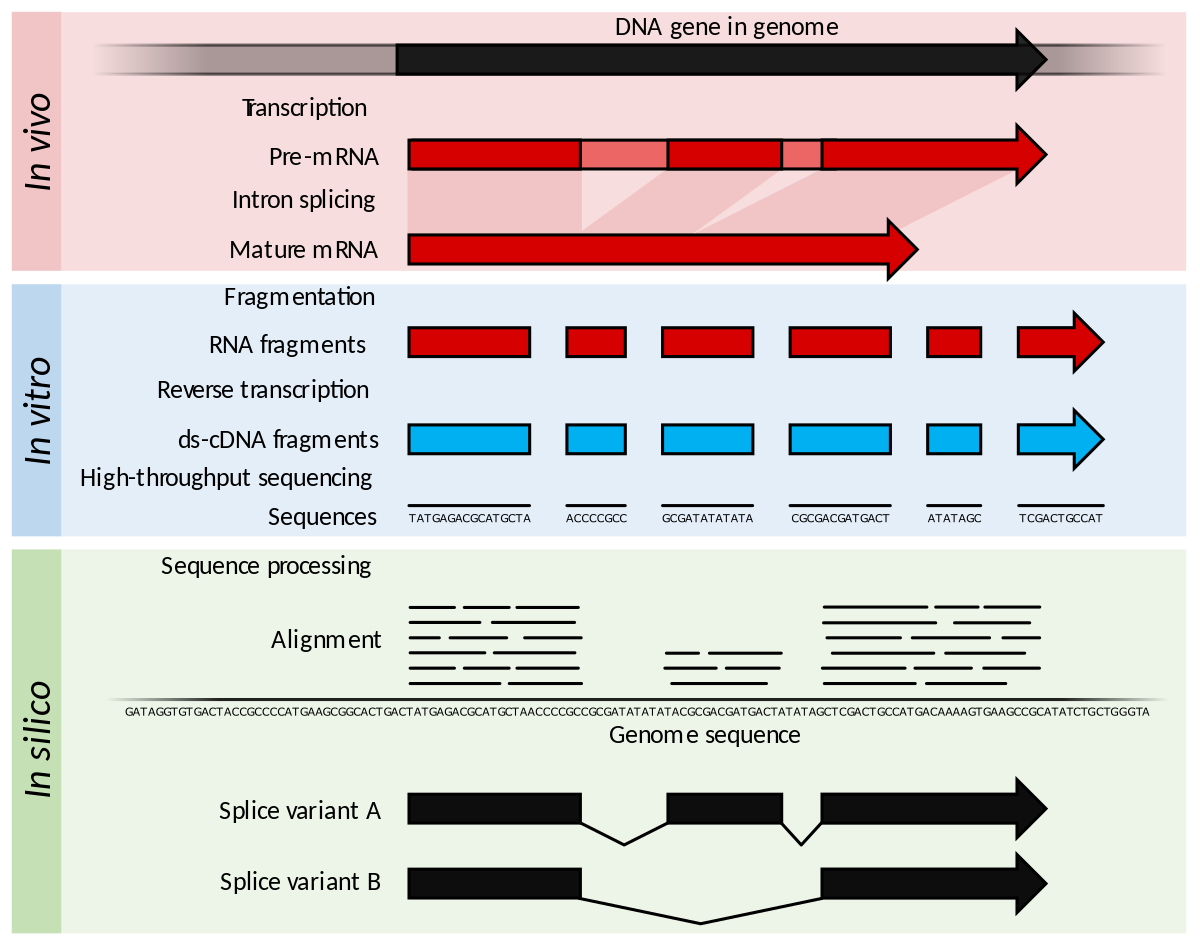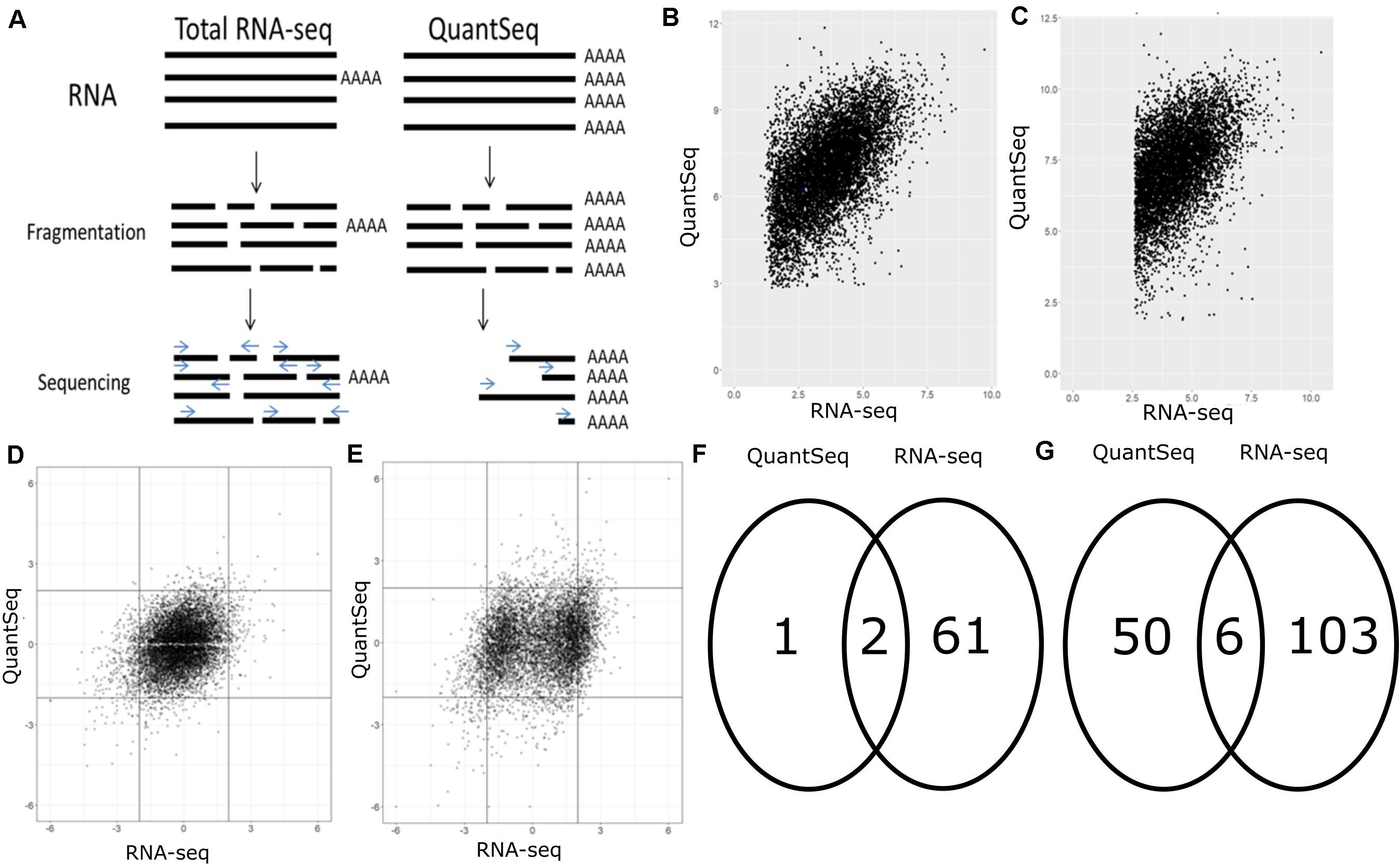
Frontiers | A Comparison of Low Read Depth QuantSeq 3′ Sequencing to Total RNA-Seq in FUS Mutant Mice

Recommendations for accurate genotyping of SARS-CoV-2 using amplicon-based sequencing of clinical samples - ScienceDirect
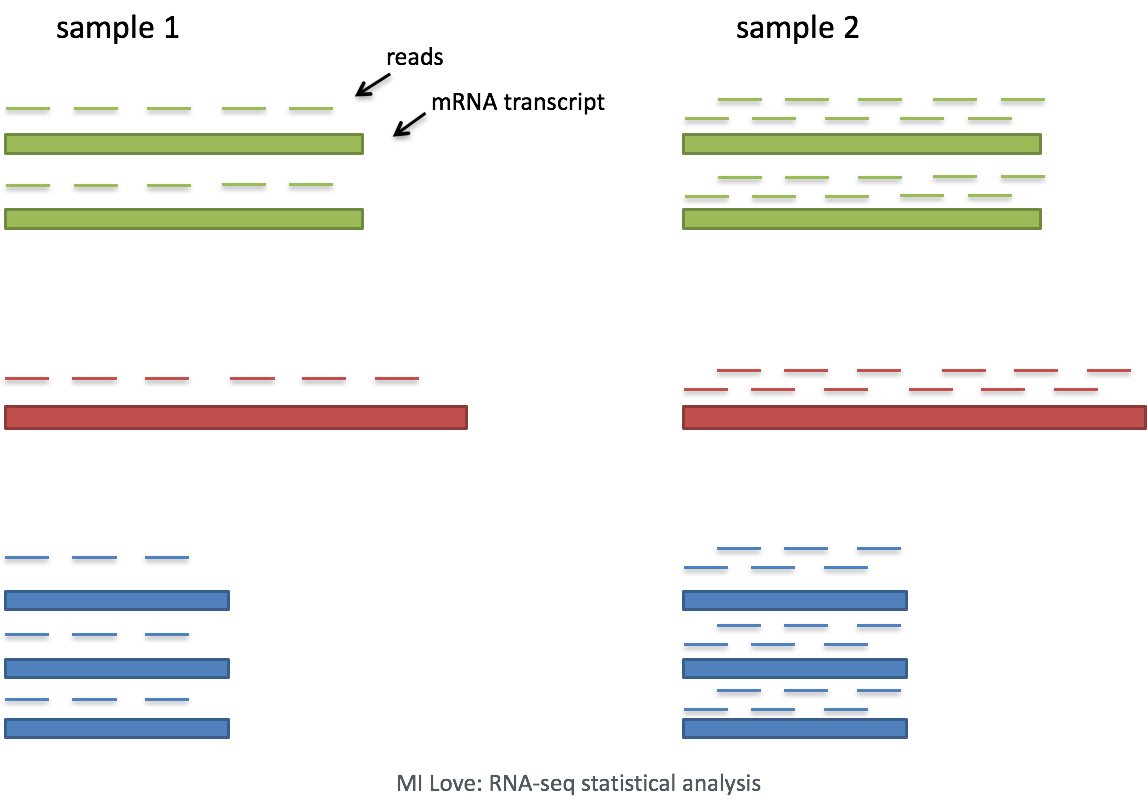
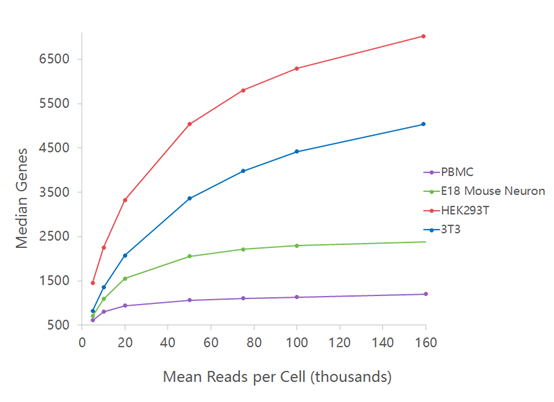
Single-cell RNA-seq: Normalization, identification of most variable genes | Introduction to single-cell RNA-seq
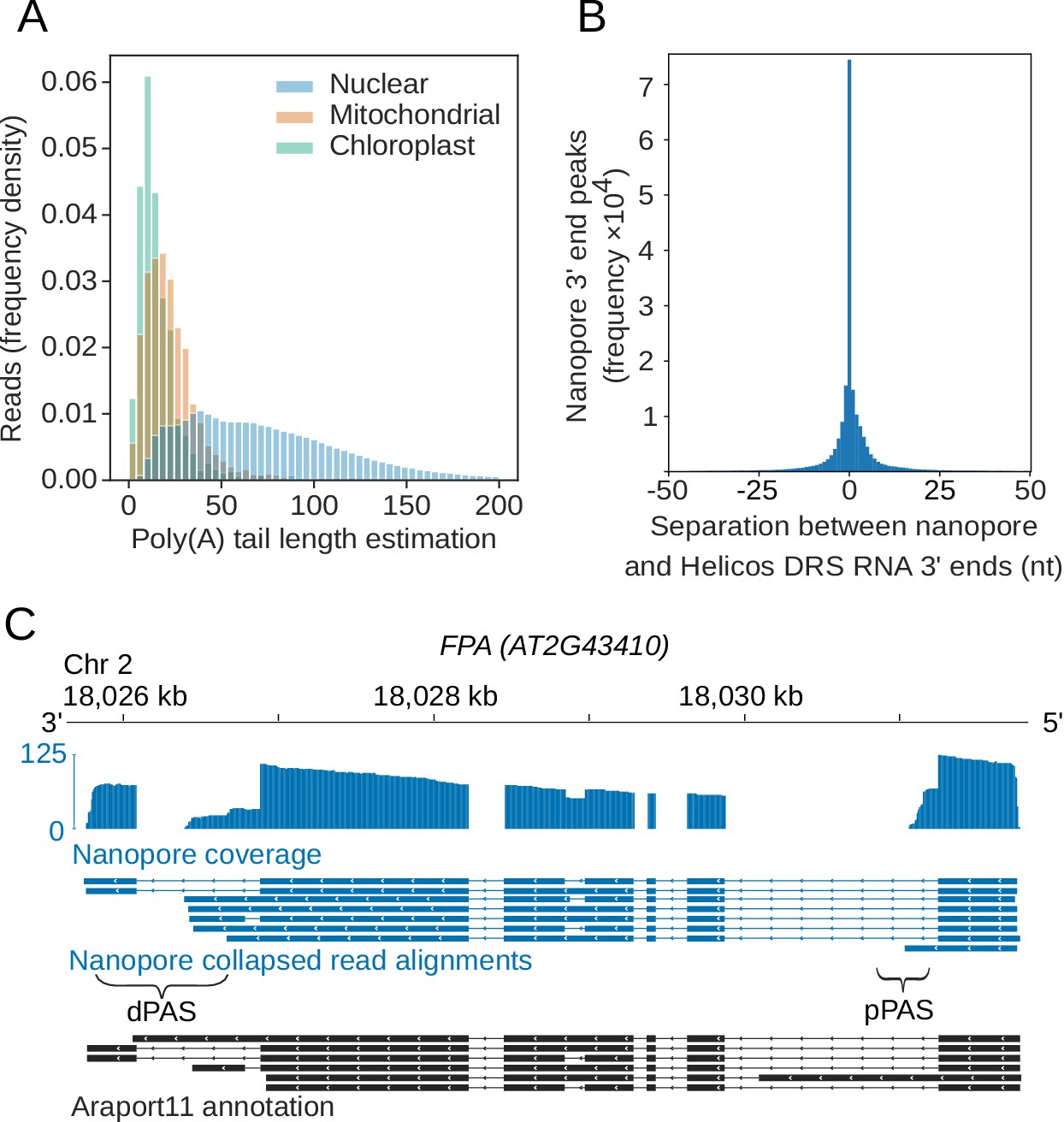
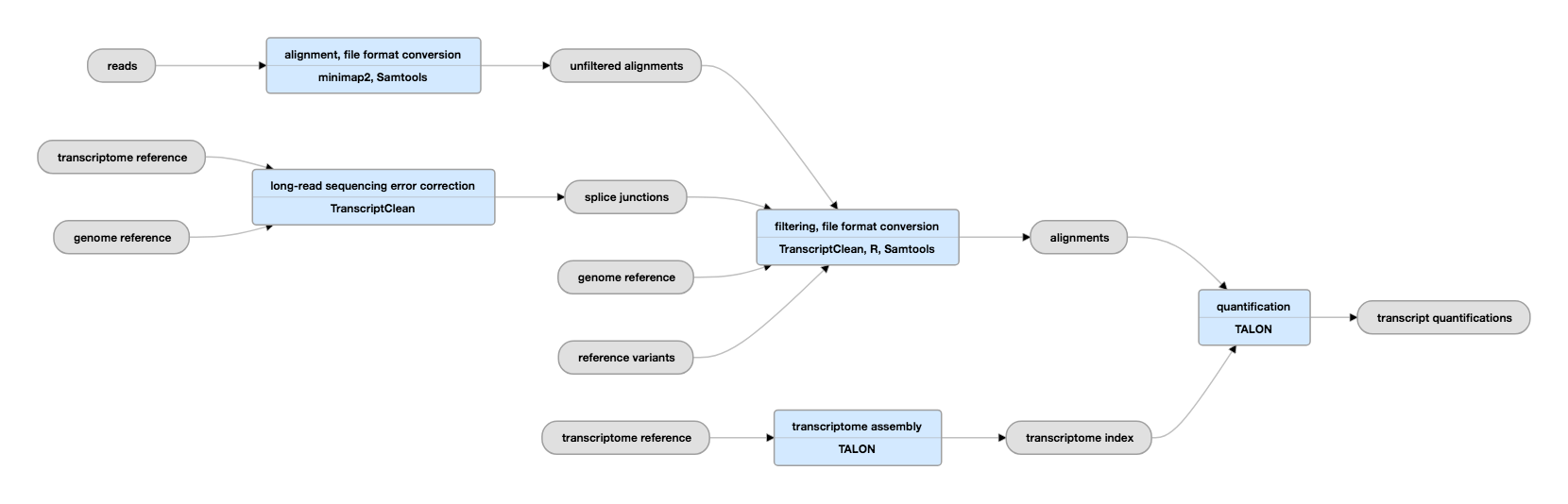
Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification | eLife
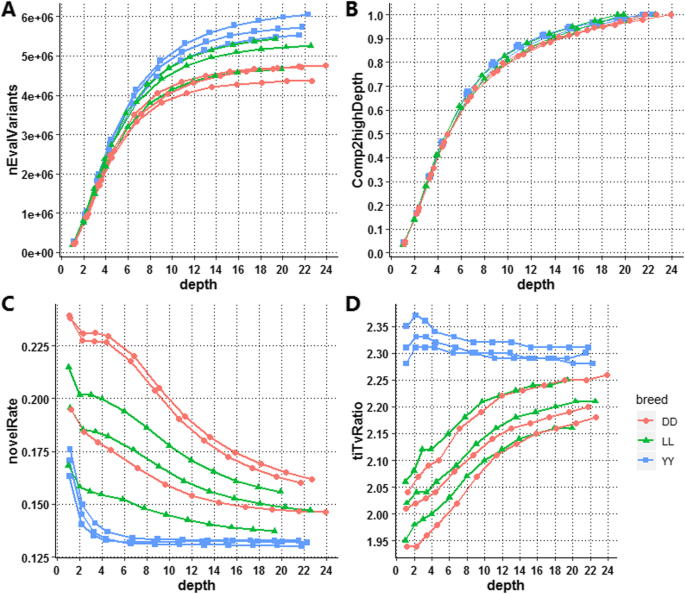
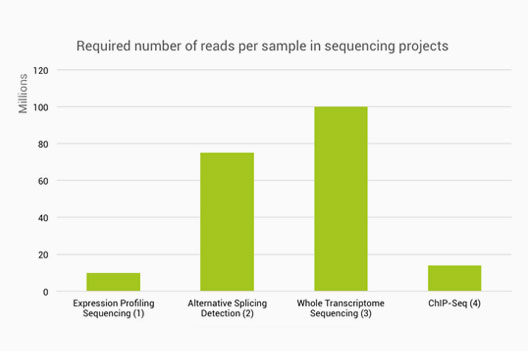
Optimal sequencing depth design for whole genome re-sequencing in pigs | BMC Bioinformatics | Full Text