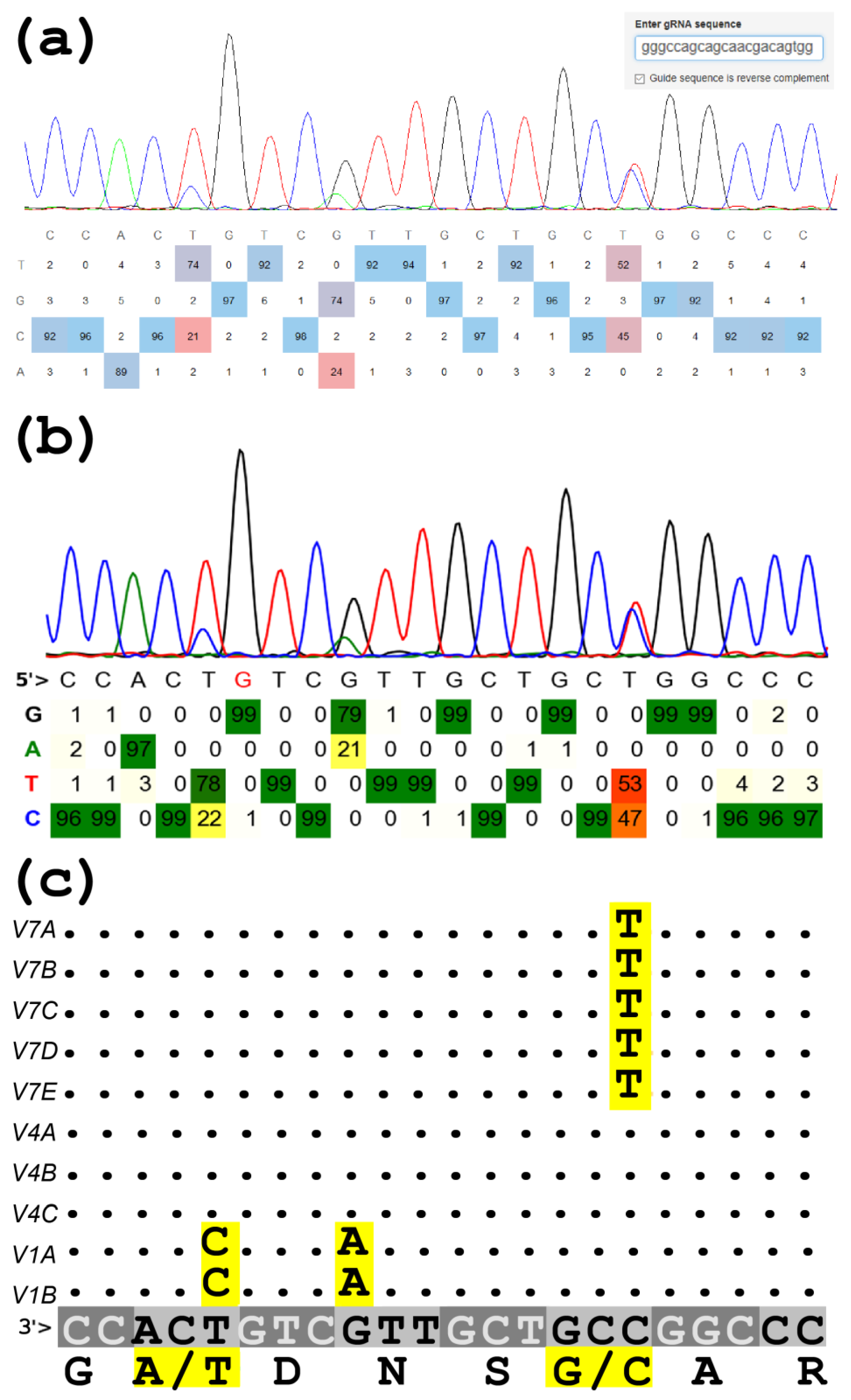
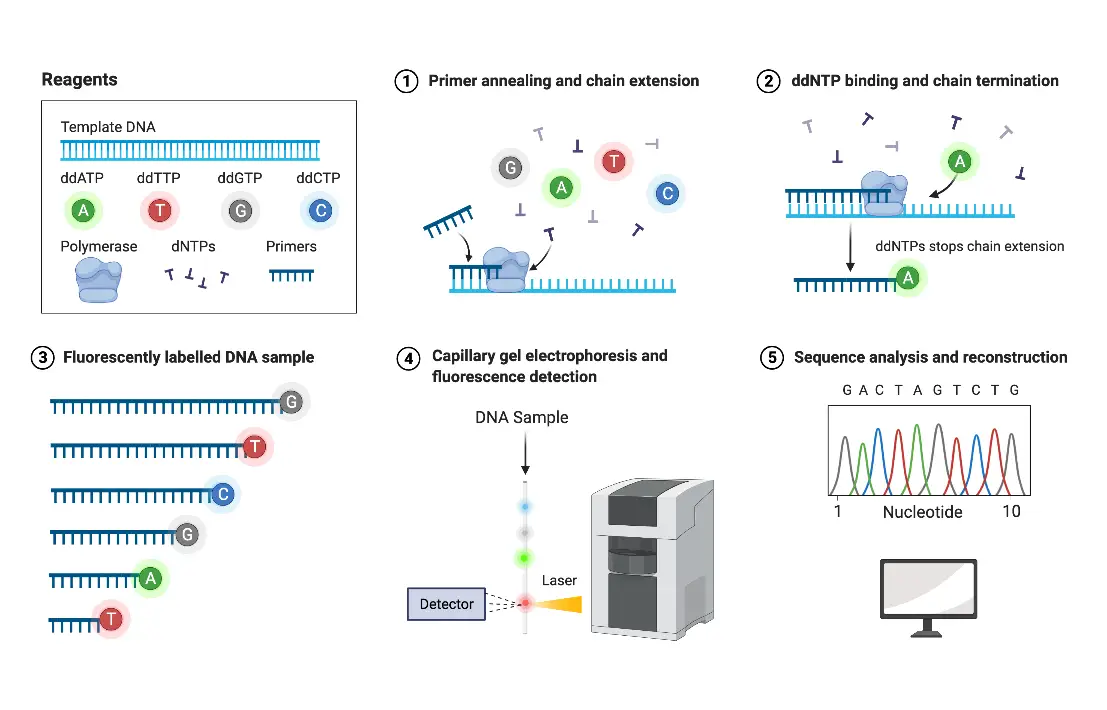
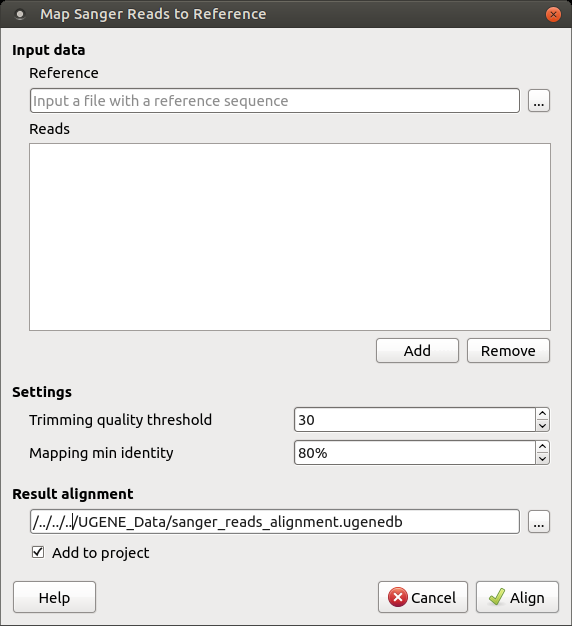
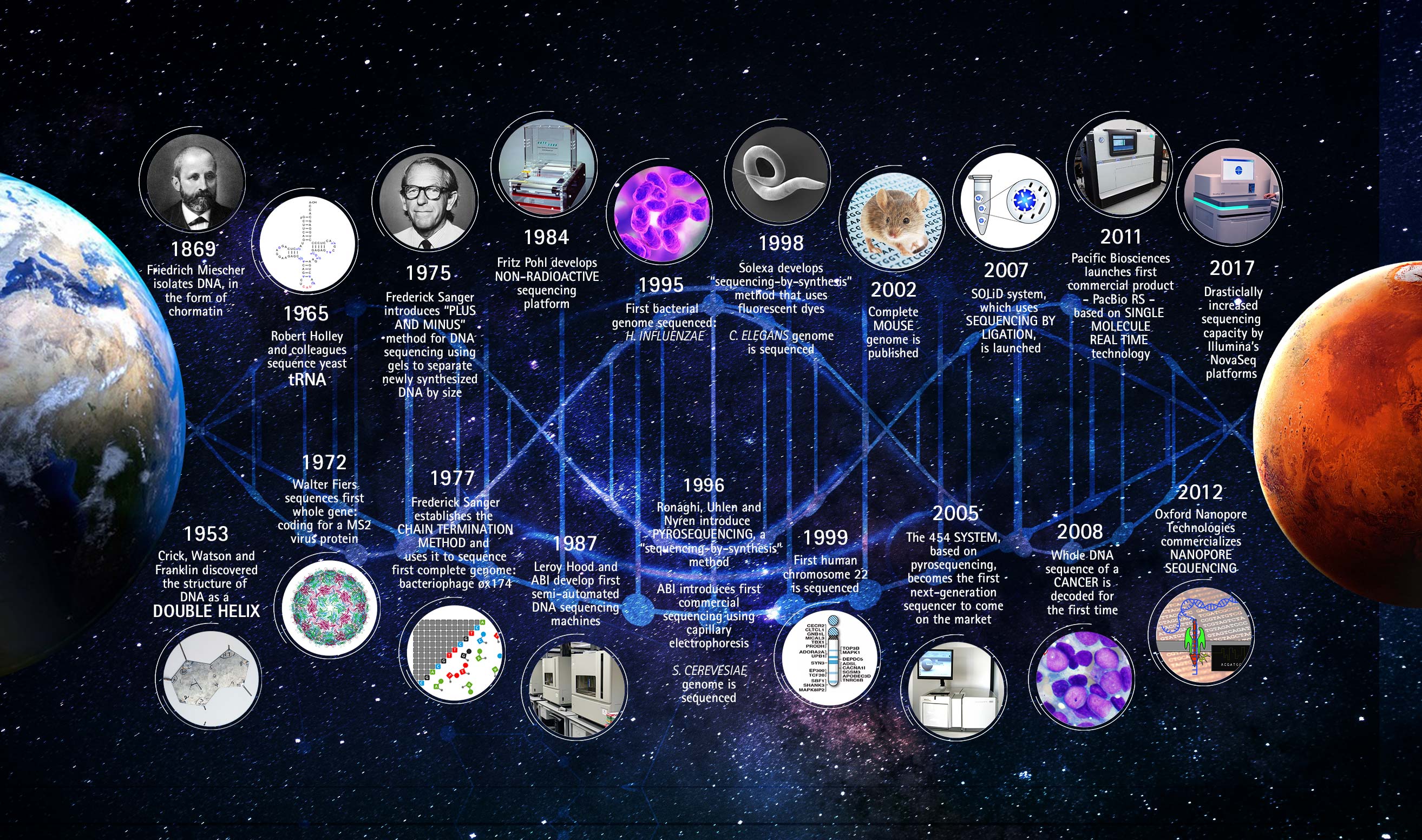
TraceTrack, an Open-Source Software for Batch Processing, Alignment and Visualization of Sanger Sequencing Chromatograms | bioRxiv

Sanger sequencing electropherogram: two mutations of NLRP7. At the top... | Download Scientific Diagram

Sanger sequencing results for T/TG track. Reference DNA T/TG track (top... | Download Scientific Diagram
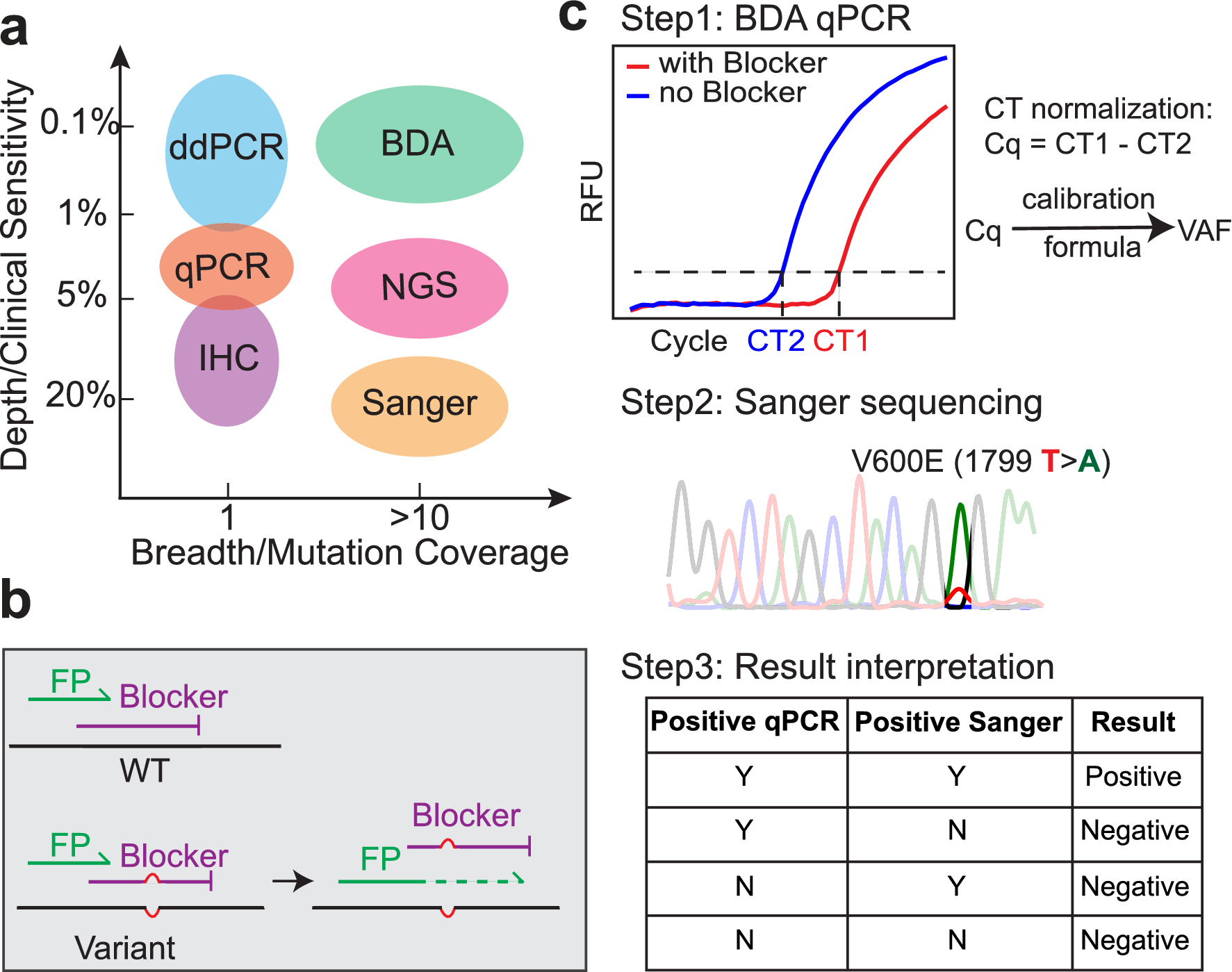
High sensitivity sanger sequencing detection of BRAF mutations in metastatic melanoma FFPE tissue specimens | Scientific Reports

MultiEditR: The first tool for the detection and quantification of RNA editing from Sanger sequencing demonstrates comparable fidelity to RNA-seq: Molecular Therapy - Nucleic Acids
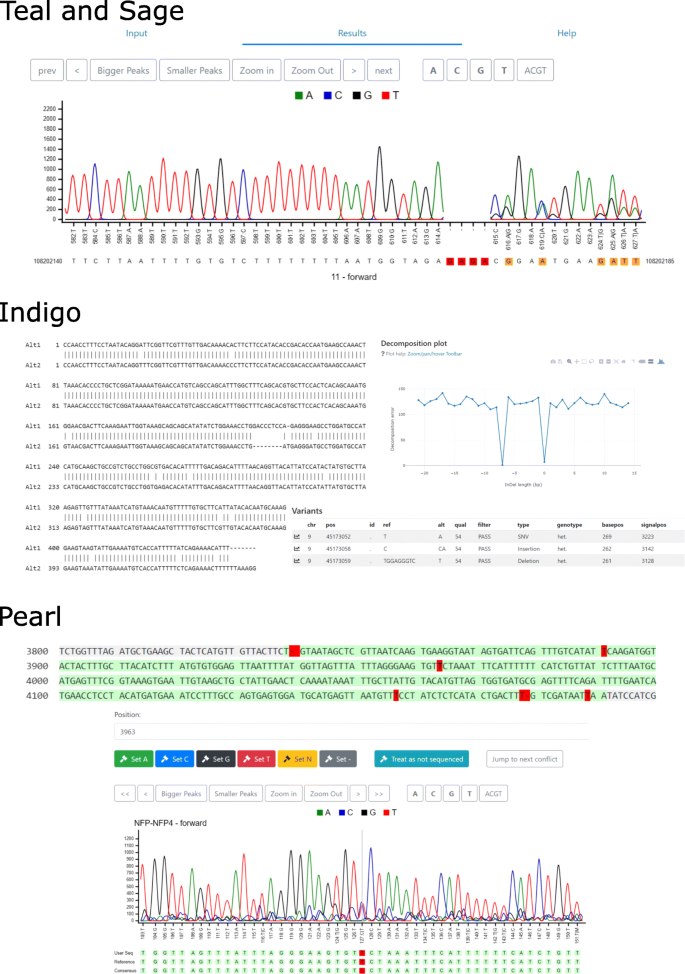
Tracy: basecalling, alignment, assembly and deconvolution of sanger chromatogram trace files | BMC Genomics | Full Text
![PDF] A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. | Semantic Scholar PDF] A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/4e241317279e23f91b4975c66bf4d5952df4aeec/1-Figure3-1.png)
![Tutorial] How to identify bacteria using a single Sanger sequence – EzBioCloud Help center Tutorial] How to identify bacteria using a single Sanger sequence – EzBioCloud Help center](https://help.ezbiocloud.net/wp-content/uploads/2018/01/ab1.png)







![Tutorial] How to identify bacteria using a single Sanger sequence – EzBioCloud Help center Tutorial] How to identify bacteria using a single Sanger sequence – EzBioCloud Help center](https://help.ezbiocloud.net/wp-content/uploads/2018/01/ezeditor2_case1_4.png)