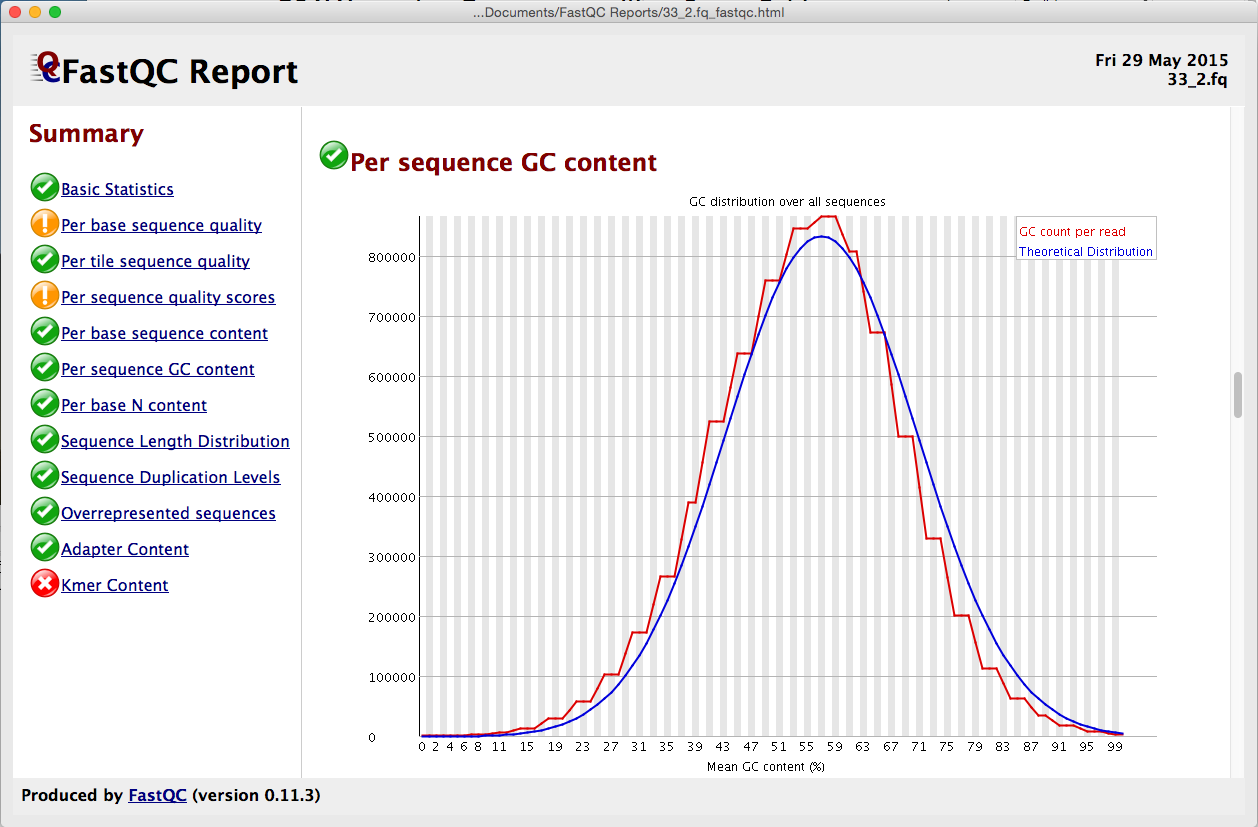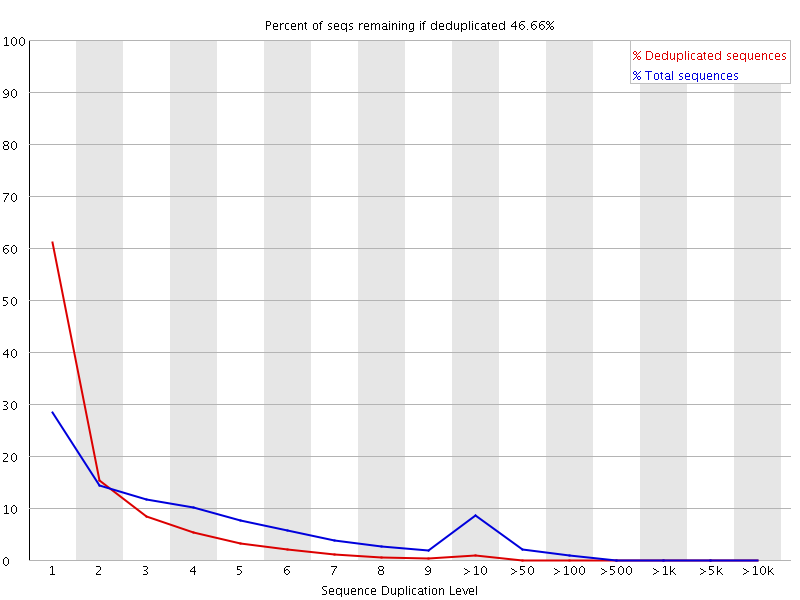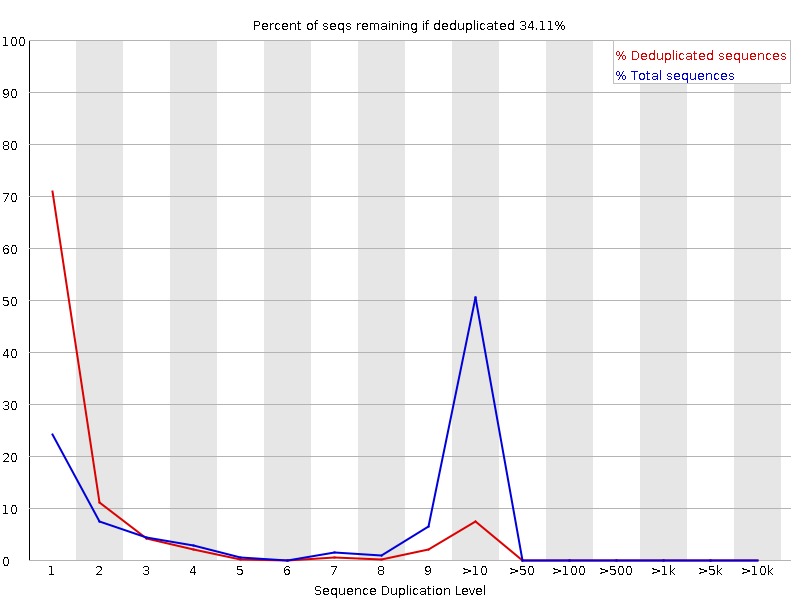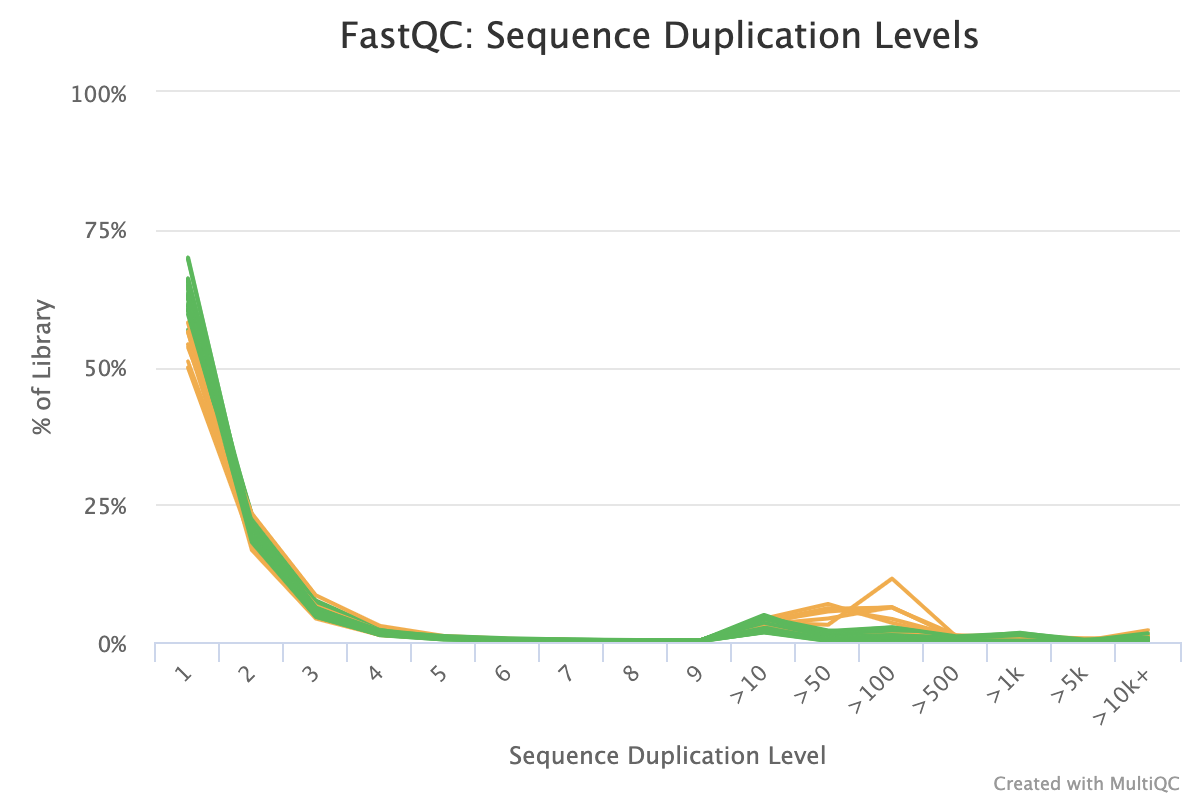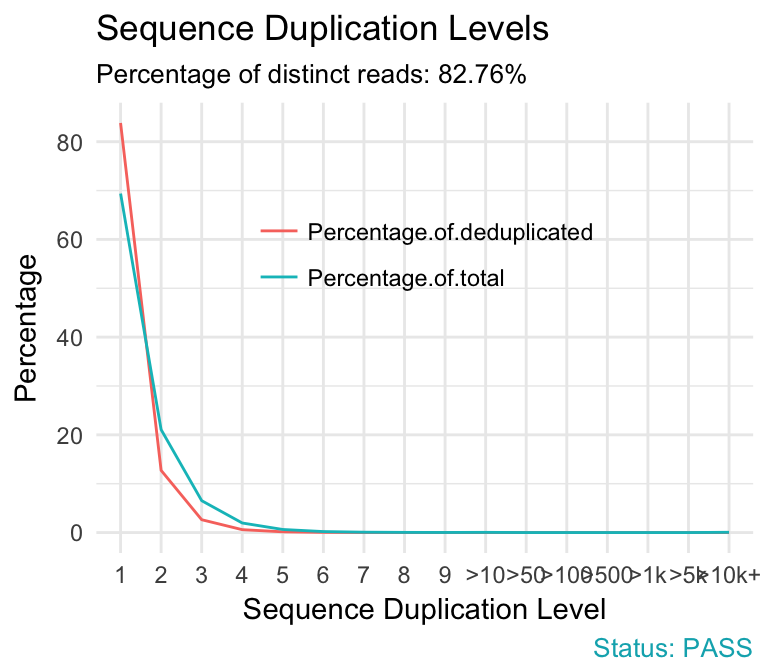
fastqcr: An R Package Facilitating Quality Controls of Sequencing Data for Large Numbers of Samples | R-bloggers
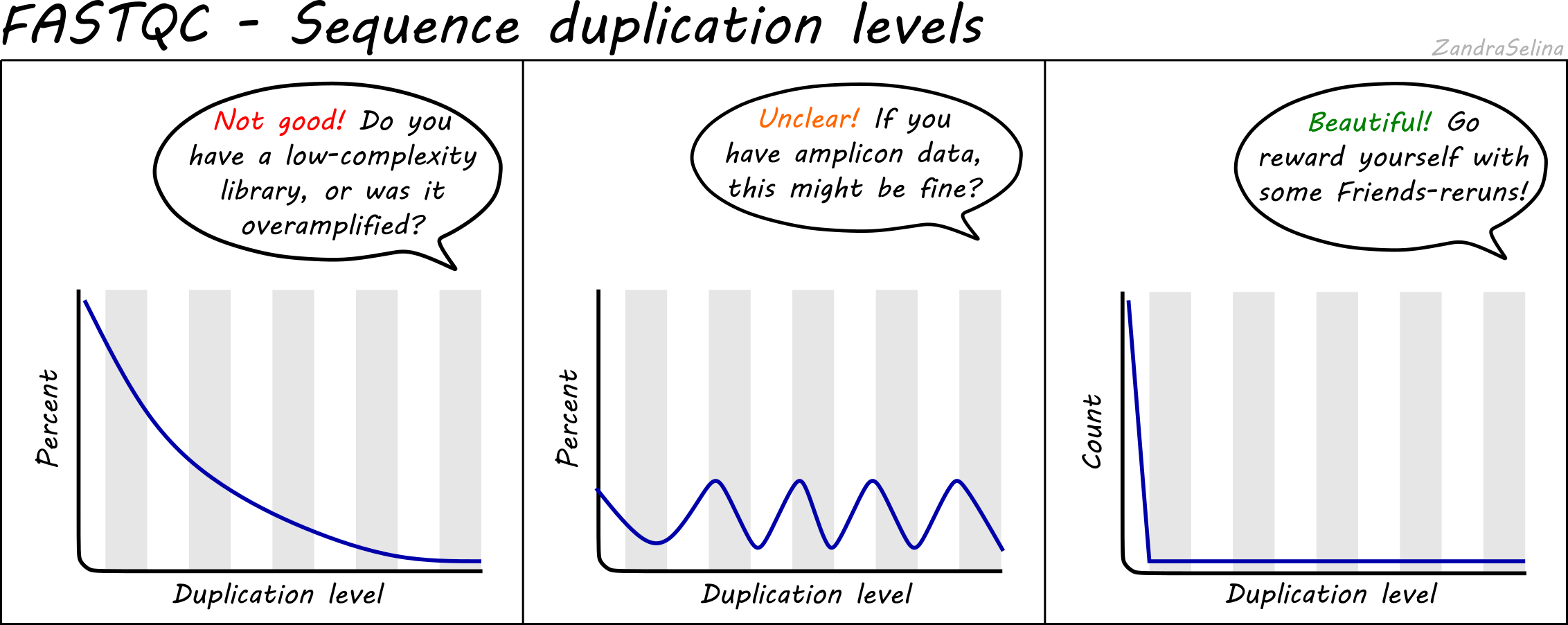
Why does FASTQC show unexpectedly high sequence duplication levels (PCR-duplicates)? | DNA Technologies Core

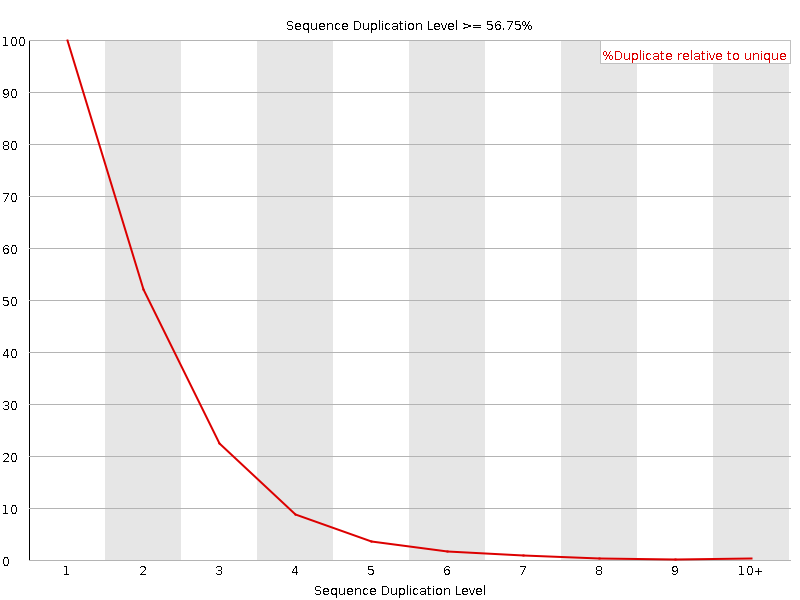
FASTQ Quality Assurance Tools - Bioinformatics Team (BioITeam) at the University of Texas - UT Austin Wikis
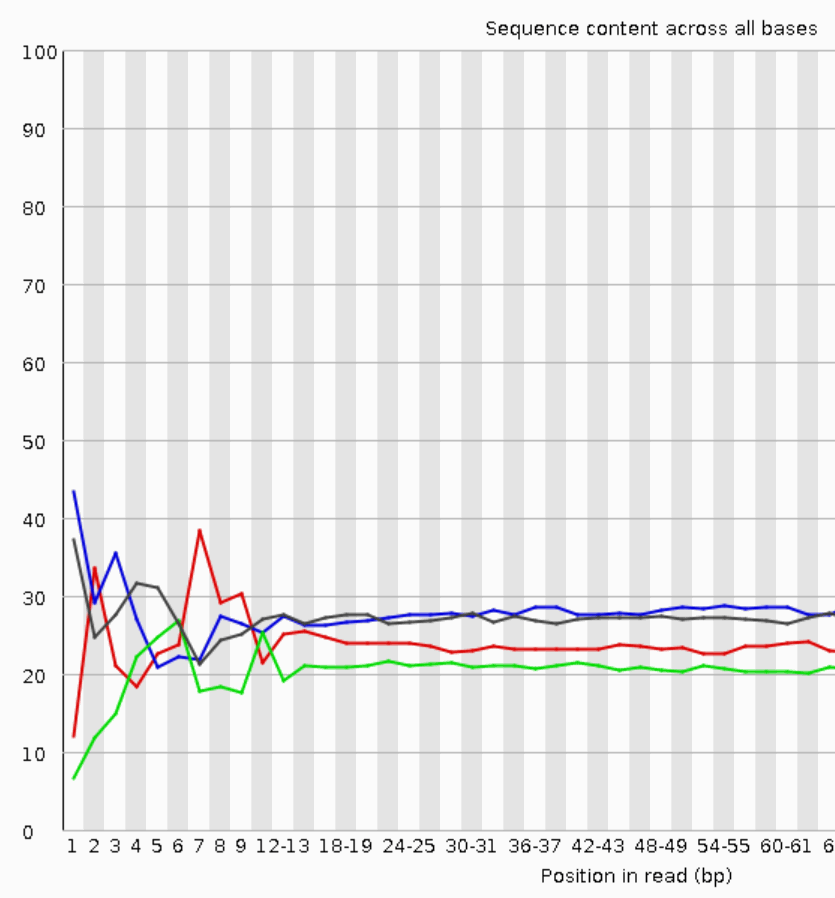
Quality control: Assessing FASTQC results | Introduction to RNA-Seq using high-performance computing - ARCHIVED
FastQC for RNA-Seq-Fails for per base seq content, gc content, duplication levels, adapter content and kmer content? | ResearchGate

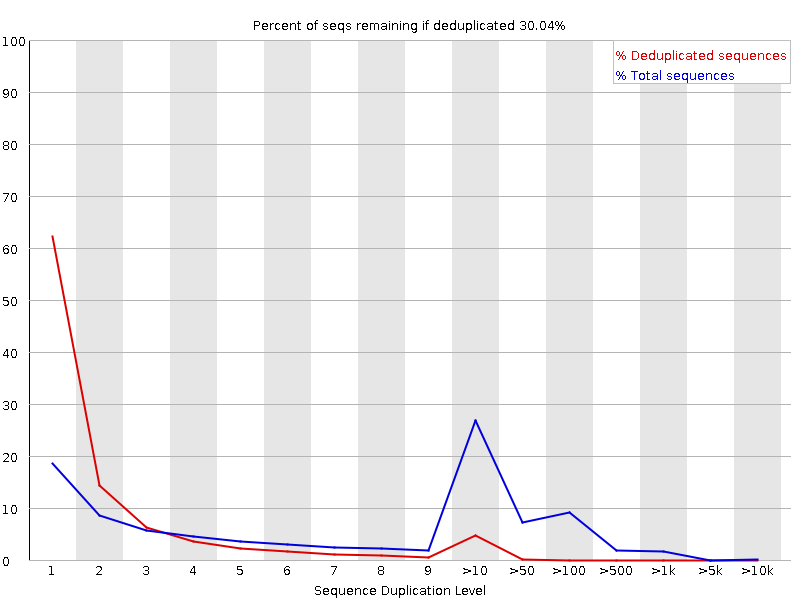
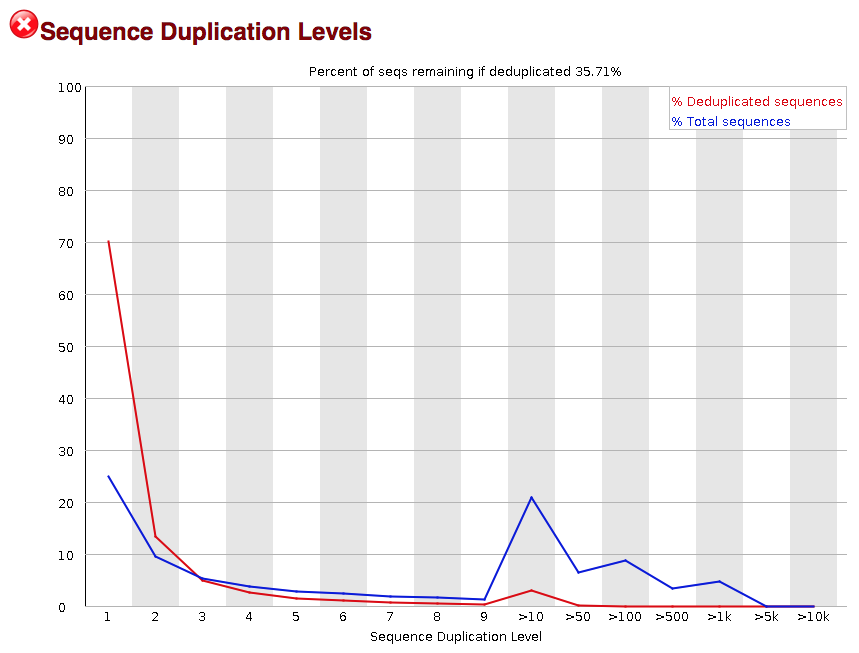
SciBear🇺🇦 on Twitter: "(6/7) Sequence duplication levels. This module counts the degree of #duplication for every sequence in the set and creates a plot showing the relative number of sequences with different