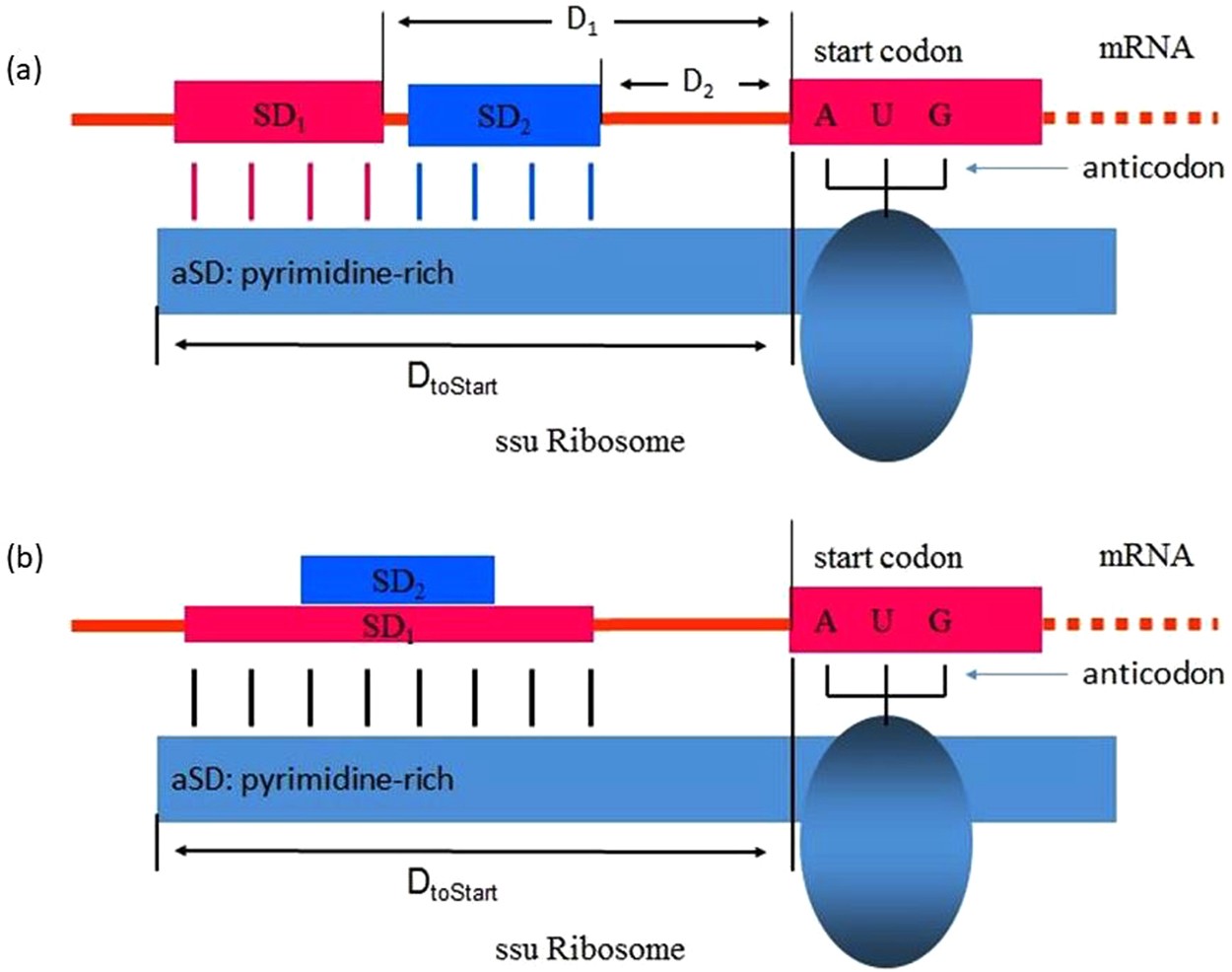
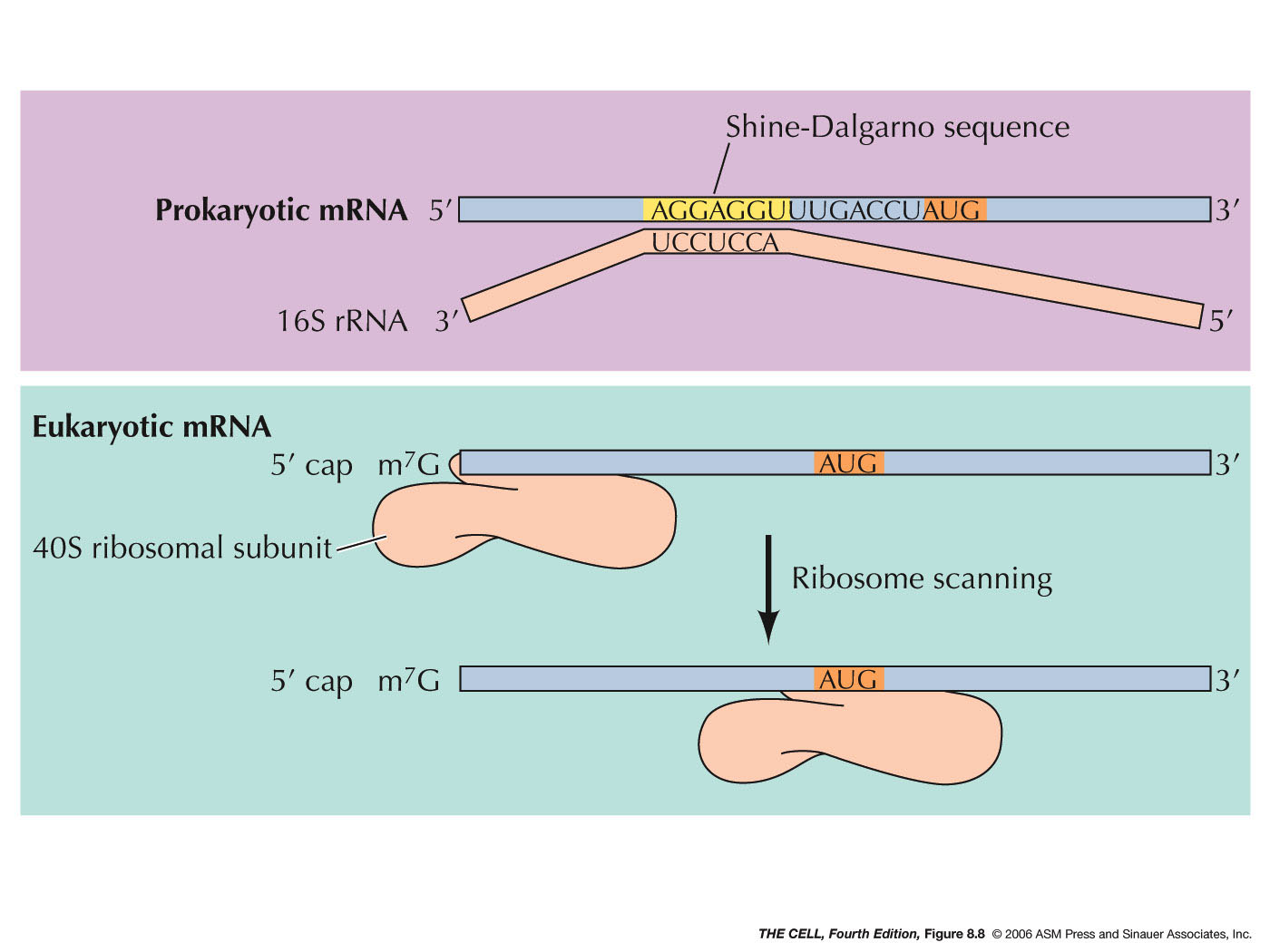
![PDF] Shine-Dalgarno Sequences Play an Essential Role in the Translation of Plastid mRNAs in Tobacco | Semantic Scholar PDF] Shine-Dalgarno Sequences Play an Essential Role in the Translation of Plastid mRNAs in Tobacco | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/994e2f8be82562f98a5c4bc645df7d19baf22bfb/2-Figure1-1.png)
PDF] Shine-Dalgarno Sequences Play an Essential Role in the Translation of Plastid mRNAs in Tobacco | Semantic Scholar

An extended Shine–Dalgarno sequence in mRNA functionally bypasses a vital defect in initiator tRNA | PNAS

Leveraging genome-wide datasets to quantify the functional role of the anti- Shine–Dalgarno sequence in regulating translation efficiency | Open Biology
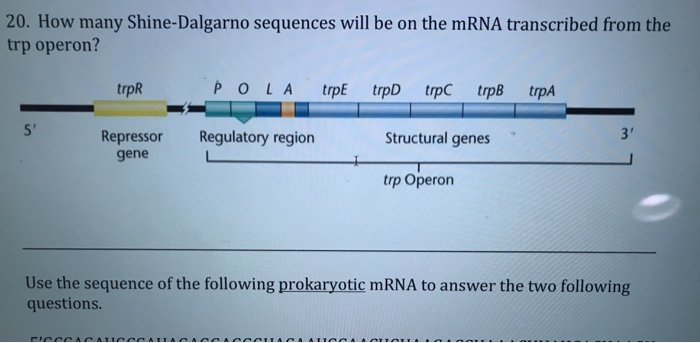
In prokaryotes, the ribosomes binding site on mRNA is called(a)Hogness sequence(b)Shine- Dalgarno sequence(c)Prinbow sequence(d)TATA- box
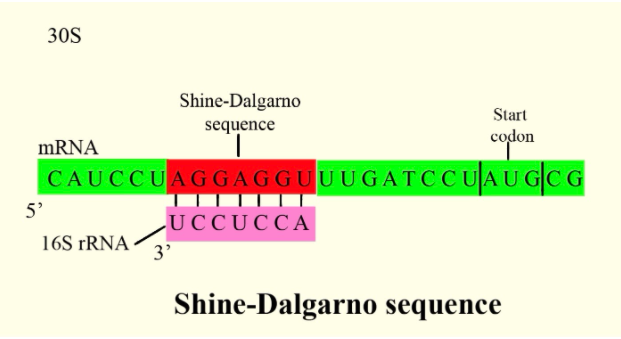
Translation initiation region sequence preferences in Escherichia coli | BMC Molecular Biology | Full Text

What is the Difference Between Shine Dalgarno and Kozak Sequence | Compare the Difference Between Similar Terms
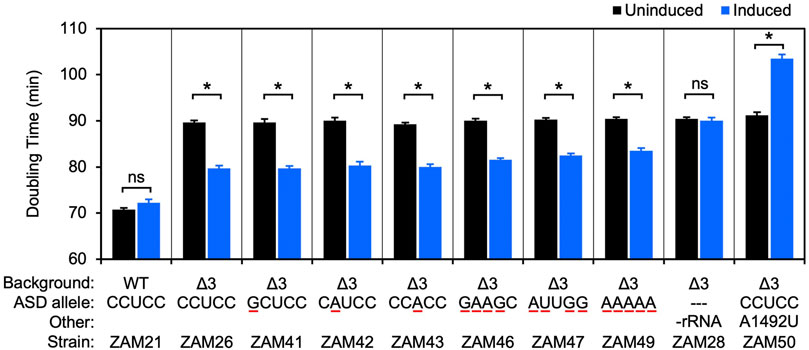
Frontiers | Comparative Analysis of anti-Shine- Dalgarno Function in Flavobacterium johnsoniae and Escherichia coli
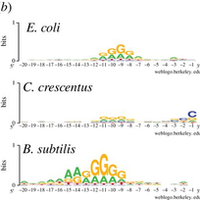
Leveraging genome-wide datasets to quantify the functional role of the anti- Shine–Dalgarno sequence in regulating translation efficiency - Publications - Amaral Lab
![PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/ca6e3ce301334ffc5d3a7df6b8a8e5d2bc19ed16/84-Table3.3-1.png)
PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar
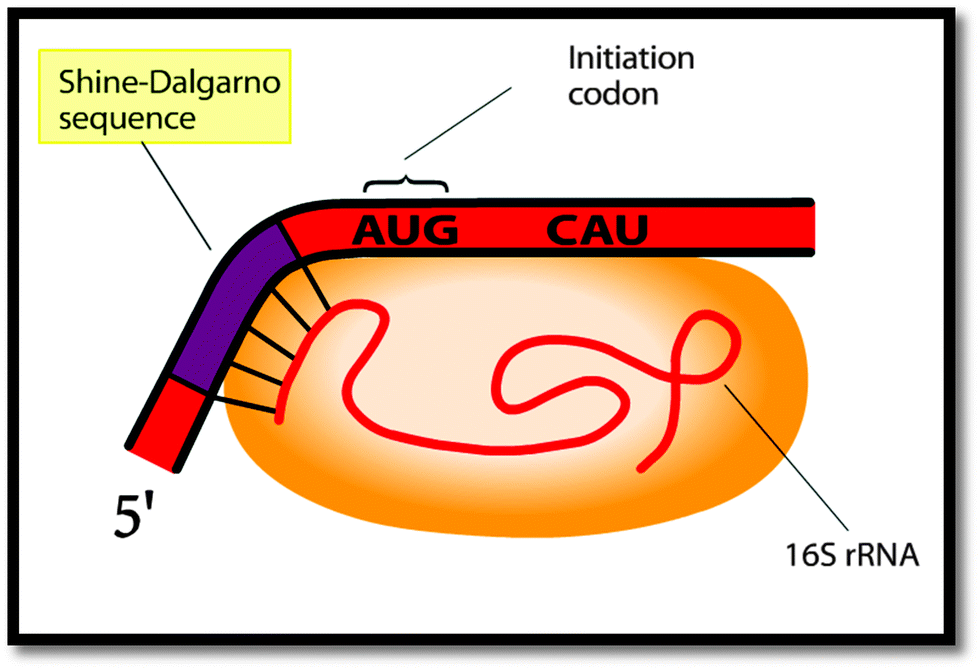
What do biochemistry students pay attention to in external representations of protein translation? The case of the Shine–Dalgarno sequence - Chemistry Education Research and Practice (RSC Publishing) DOI:10.1039/C5RP00001G

Defining the anti-Shine-Dalgarno sequence interaction and quantifying its functional role in regulating translation efficiency | bioRxiv

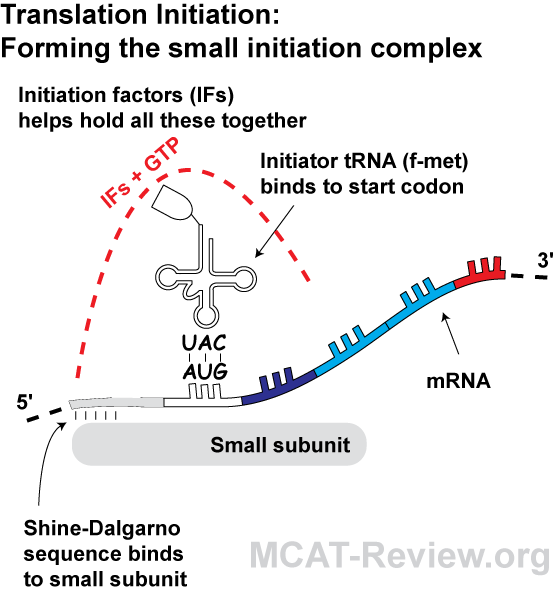
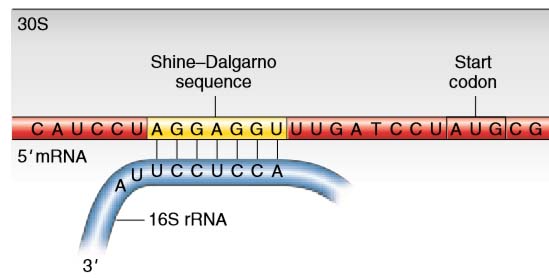
Shine-Dalgarno Motif Ribosome binding site located about 13 bases upstream of AUG start codon SD sequence is: 5'-AGGAGGU-3' Middle GGAG is more highly. - ppt download

Accessibility of the Shine-Dalgarno Sequence Dictates N-Terminal Codon Bias in E. coli - ScienceDirect