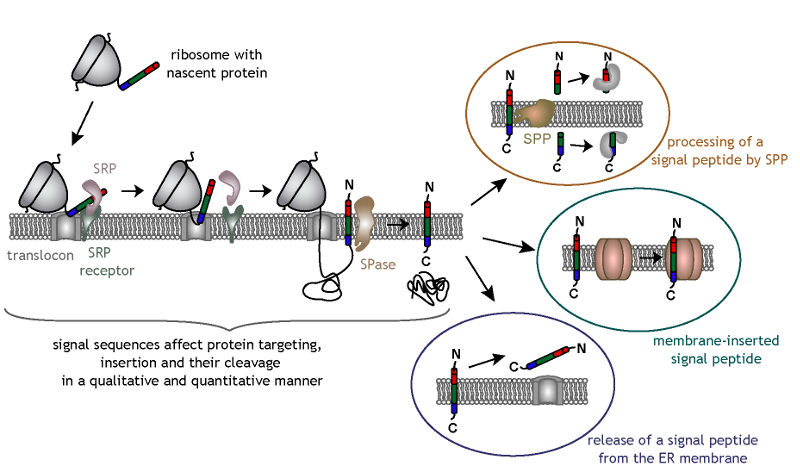
Structure and topology around the cleavage site regulate post-translational cleavage of the HIV-1 gp160 signal peptide | eLife
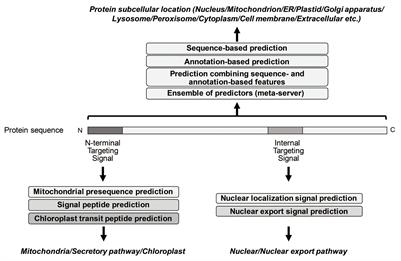
Frontiers | Tools for the Recognition of Sorting Signals and the Prediction of Subcellular Localization of Proteins From Their Amino Acid Sequences

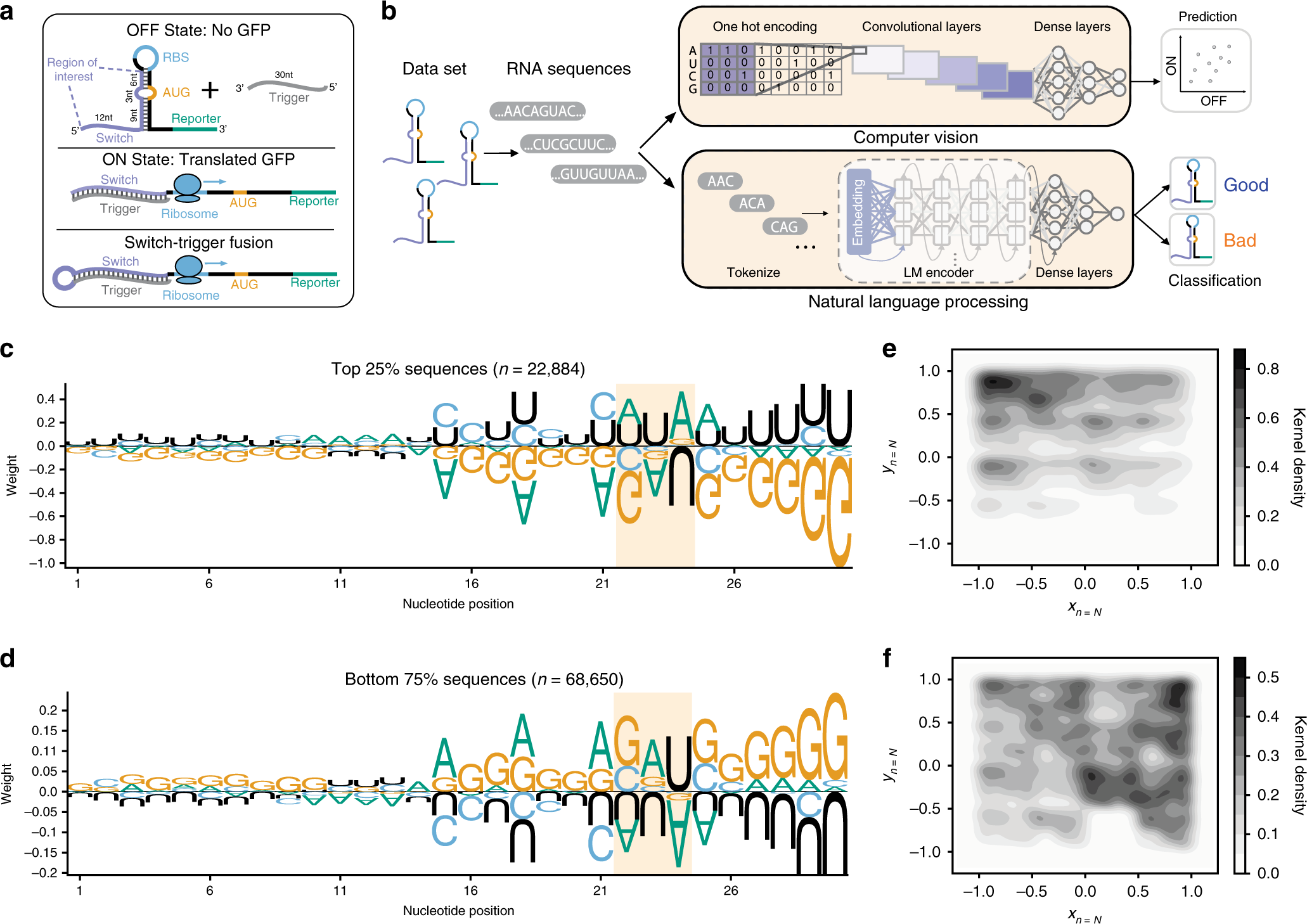
Proteome-Scale Screening to Identify High-Expression Signal Peptides with Minimal N-Terminus Biases via Yeast Display | ACS Synthetic Biology
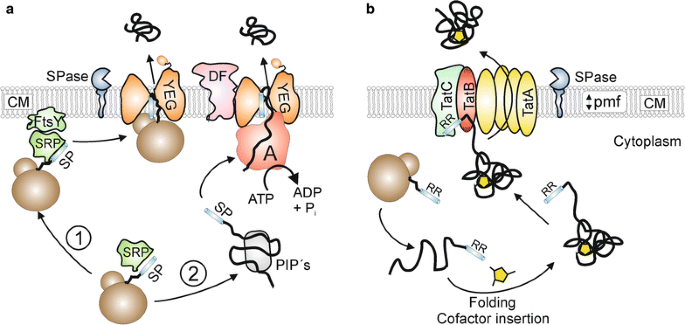
Signal peptides for recombinant protein secretion in bacterial expression systems | Microbial Cell Factories | Full Text
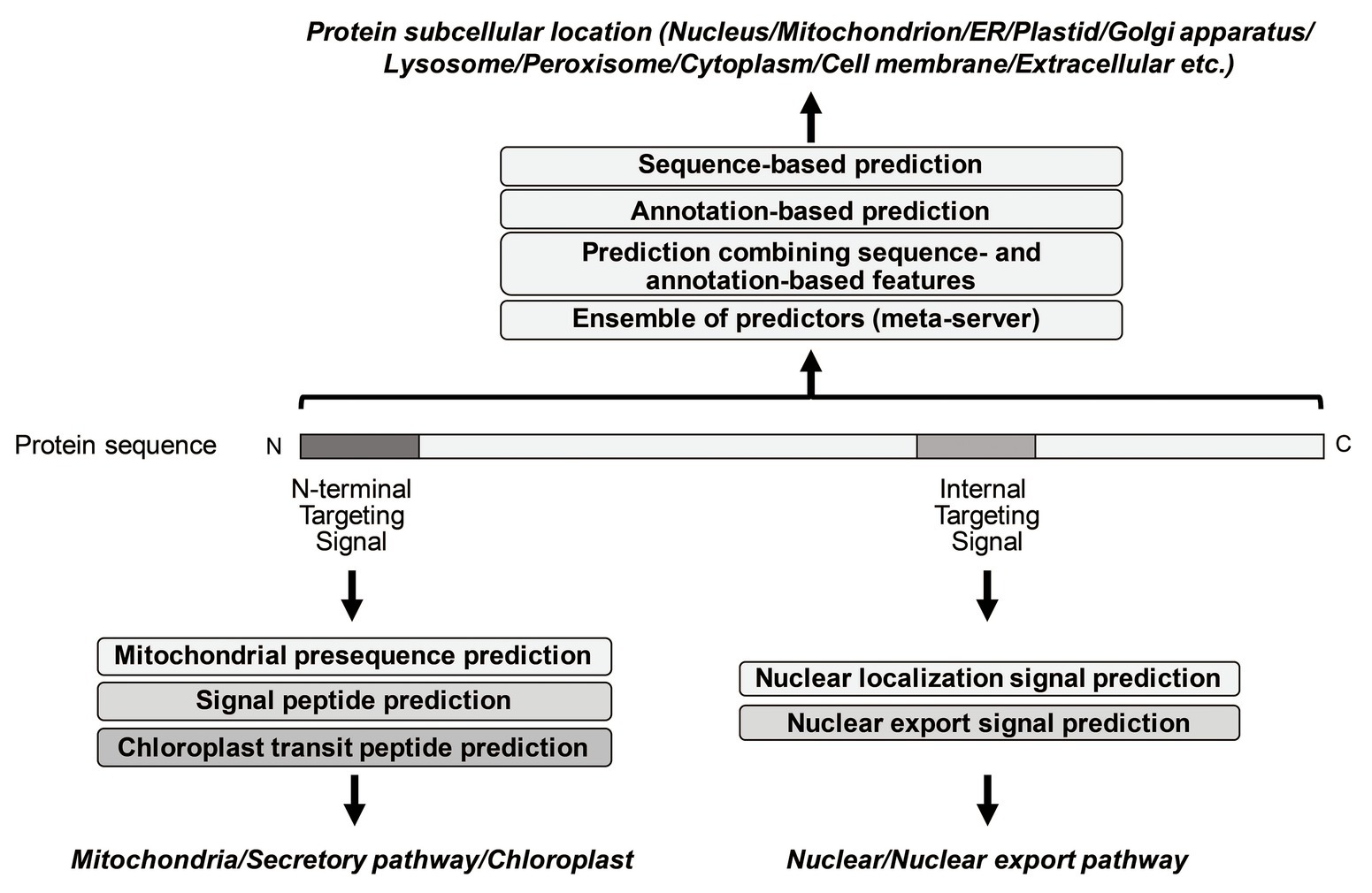
Frontiers | Tools for the Recognition of Sorting Signals and the Prediction of Subcellular Localization of Proteins From Their Amino Acid Sequences

SignalP 6.0 achieves signal peptide prediction across all types using protein language models | bioRxiv

Signal peptide prediction. Signal peptide sequence was predicted using... | Download Scientific Diagram

SignalP 6.0 predicts all five types of signal peptides using protein language models | Nature Biotechnology
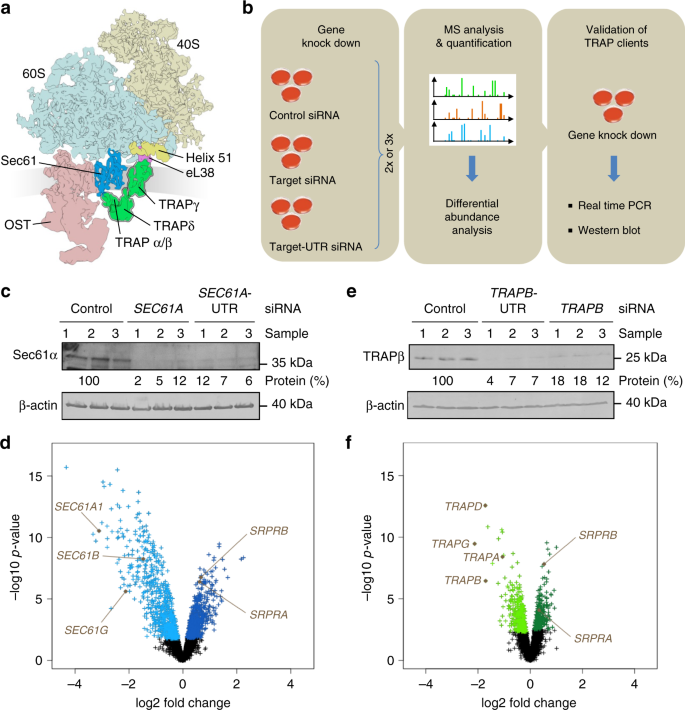
Proteomics reveals signal peptide features determining the client specificity in human TRAP-dependent ER protein import | Nature Communications
![Nuclear export signal (NES) prediction of candidate proteins based on the NetNES 1.1 prediction tool [16]. Nuclear export signal (NES) prediction of candidate proteins based on the NetNES 1.1 prediction tool [16].](https://s3-eu-west-1.amazonaws.com/ppreviews-plos-725668748/1781378/preview.jpg)
Nuclear export signal (NES) prediction of candidate proteins based on the NetNES 1.1 prediction tool [16].
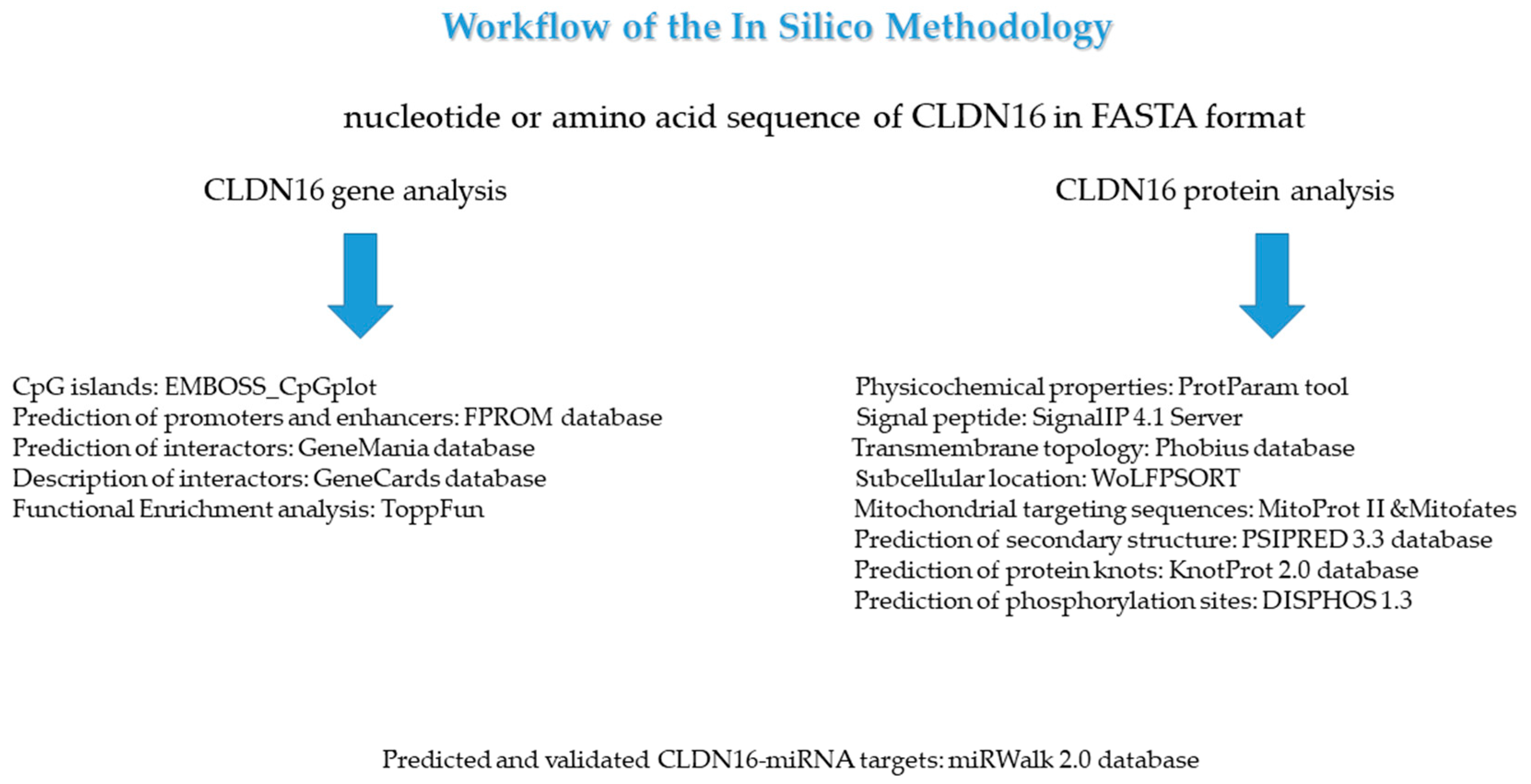
Medicina | Free Full-Text | In-Depth Bioinformatic Study of the CLDN16 Gene and Protein: Prediction of Subcellular Localization to Mitochondria
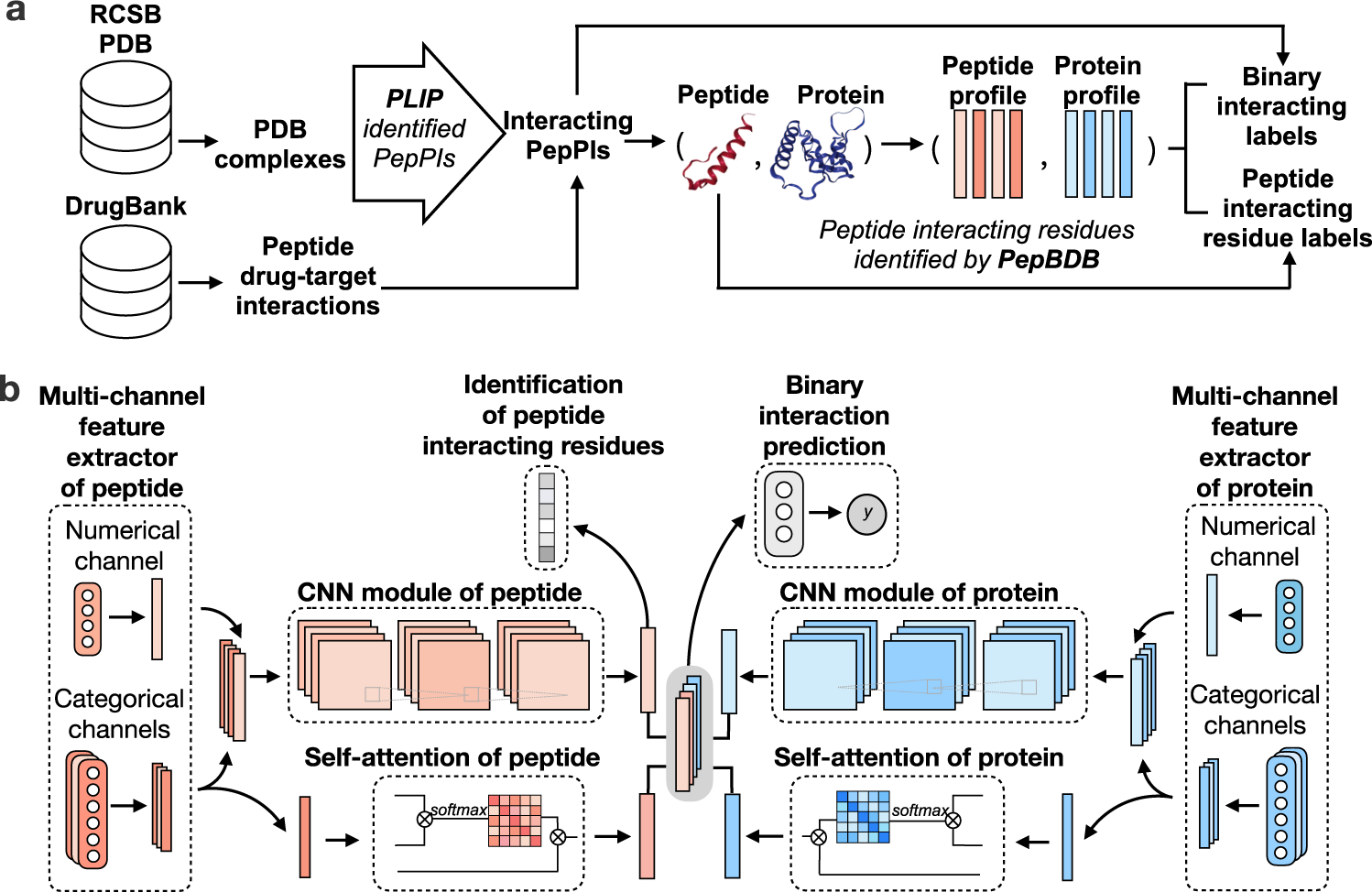


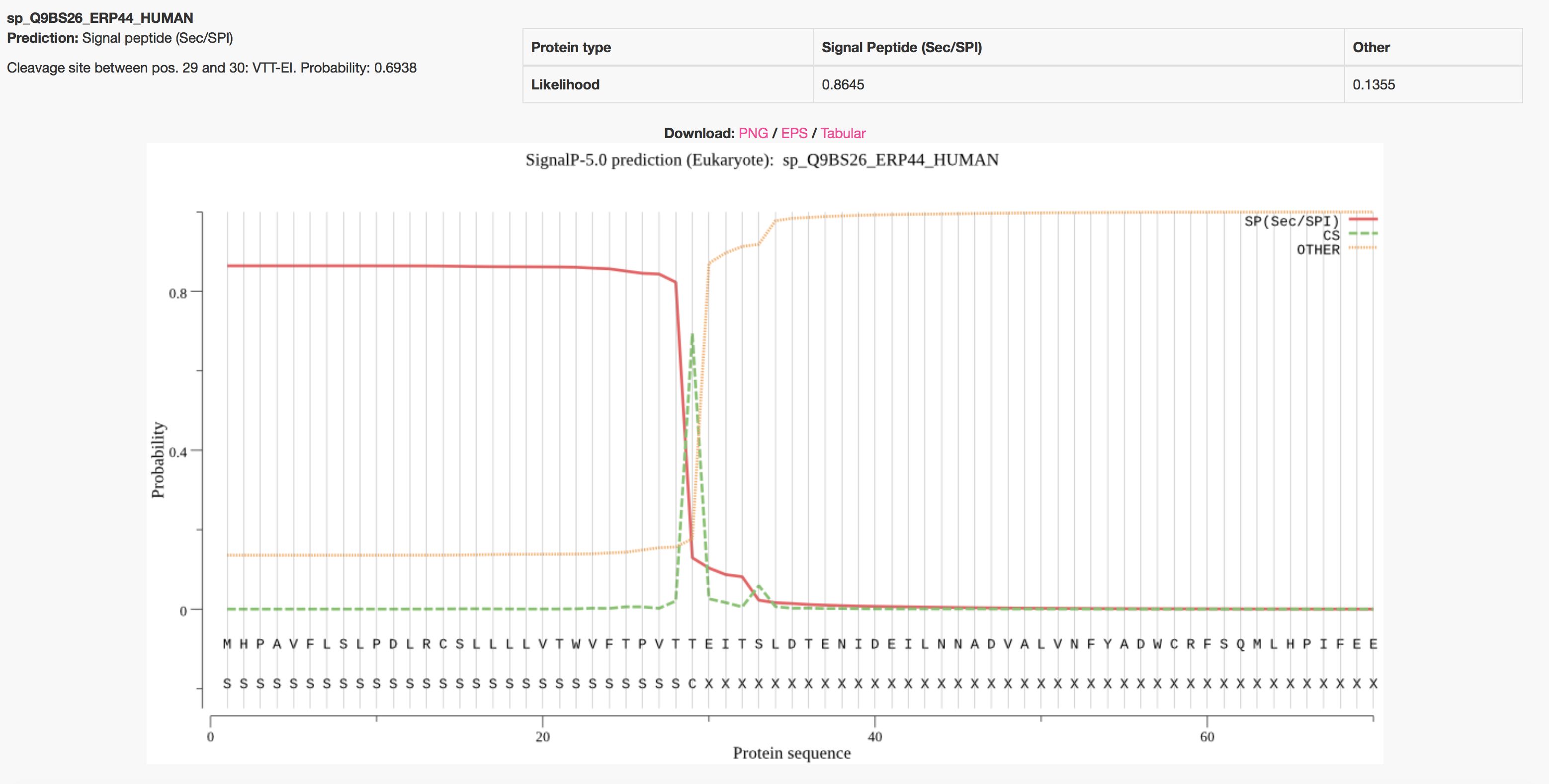

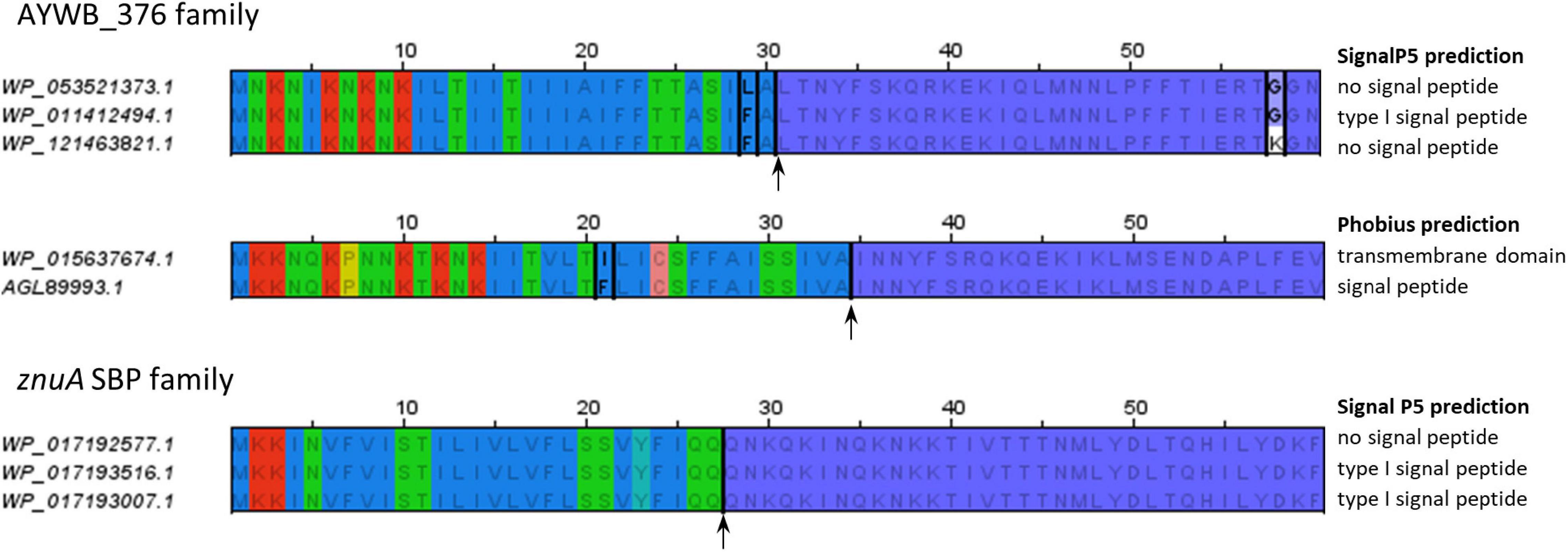


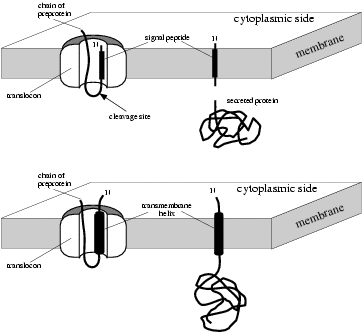
![PDF] PrediSi: prediction of signal peptides and their cleavage positions | Semantic Scholar PDF] PrediSi: prediction of signal peptides and their cleavage positions | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/296e275b6aae81a508d925835d6617c499fbe0ec/4-Figure2-1.png)